
Group members
Professor Dean Nizetic - Principal Investigator
Dr Jürgen Groet - Lecturer & Senior Research Fellow
Dr Pollyanna Goh - Post-doctoral Research Associate
Background
Cells from people with DS show in vitro similarities to the pathogenic processes underlying neurobiological features of the syndrome. These include defects in neurogenic pathways, neurite-branching, mitochondrial dysfunction, Aβ-processing and cell migration. Some of these phenotypic features are additionally reproducible in mouse DS model embryonic stem cells (ESCs) and fibroblasts.
The invention of induced pluripotent stem cells (iPSCs) has generated cellular models for specific diseases, through re-programming of primary patient cells into a state emulating human embryonic stem cells (hESC). Human iPSC (hiPSC) have created disease-in-a-dish (DIAD) systems with potential to discover new molecular mechanisms and targets relevant for pathogenesis and develop "personalised" therapeutics for specific patient subsets. DIAD-hiPSC systems have been recently developed for neurological diseases, including specific genetic subtypes of Parkinson's Disease, ALS, SMA, neuropsychiatric disorders, as well as premature ageing and mental retardation syndromes.
Specific cellular phenotypes have been reported as disease-specific pathogenetic correlates, by neural differentiation of hiPSCs and morphological/functional analysis of derived neurons. Moreover, chemical modulation of cellular phenotypes has been selectively achieved using known therapeutic drugs. hiPSC for DS have been generated alongside those for several diseases, but no specific cellular phenotypes have yet been described. These cells cannot address the correlation with clinical modalities of DS. Also, these cells were generated by random genomic integration of reprogramming DNA constructs, maximising chances of un-controlled variability among technical replicates, i.e., independent clones from the same re-programming experiment.
What we do
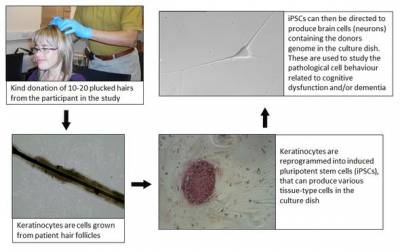
During our testing, we will ask some of our participants for a small sample of hair. These hair samples provide us with keratinocytes (the type of cells that hair follicles are) that we can convert in the lab into induced pluripotent stem cells (iPSCs).
Our work aims to characterise neurons derived from iPSCs in culture to look for differences in characteristics between the cells of our participants. The hair samples will allow us to investigate the genetic, cellular and molecular correlates of specific cognitive profiles. The genetic analysis and production of iPSCs will be undertaken by members of the research team based at UCL and Queen Mary, University of London (QMUL) respectively.

For the hair sample, we will need to pluck 6-10 hairs (including the follicle) from partipants' scalps. Hairs will be plucked at the same time to minimise any pain felt and then placed in a sealed tissue culture bottle. This will allow the our team to derive primary keratinocytes in order to perform cellular and molecular analyses, and enable the generation of iPSCs.
 Close
Close

