Wenjuan Zhang
 w.zhang@prion.ucl.ac.uk Tel: Courtauld Building, Room 101 • PubMed •
|
Research Synopsis
A prion-like mechanism is recognized as potentially underlying protein assembly in neurodegenerative diseases. The assembled proteins propagate and spread from cell-to-cell like prions (PrPSc), and exhibit prion-like conformational strains. Like mammalian prions, yeast prions exhibit “prion properties”. They propagate by a protein-only assembly process, and possess strain diversity through distinct conformers. Specifically, yeast prions can stably but reversibly exist within cell populations in soluble and high-molecular weight insoluble, aggregated forms. The protein quality control machinery involved in their propagation is highly conserved between mammals and yeast, which underlies the wide relevance of the study of yeast prions.
We aim to elucidate the structures and mechanisms of prion-like protein assemblies in neurodegenerative diseases using an integrated biochemical and structural approaches, especially cryo-electron microscopy. On the one hand, our group will work with the Alzheimer’s disease group to study the cryo-EM structures and identify the regulating factors of amyloid-β (Aβ) assemblies in Alzheimer's disease, using human brain-derived samples and mouse models. On the other hand, we will study yeast prions, in particular [URE3] and [PSI+], which are formed by assembly of Ure2p and Sup35p, respectively. Our research will shed light on how prion-like proteins are assembled and regulated, with implications for rationalized design of diagnostic tools and therapeutics for neurodegenerative diseases.
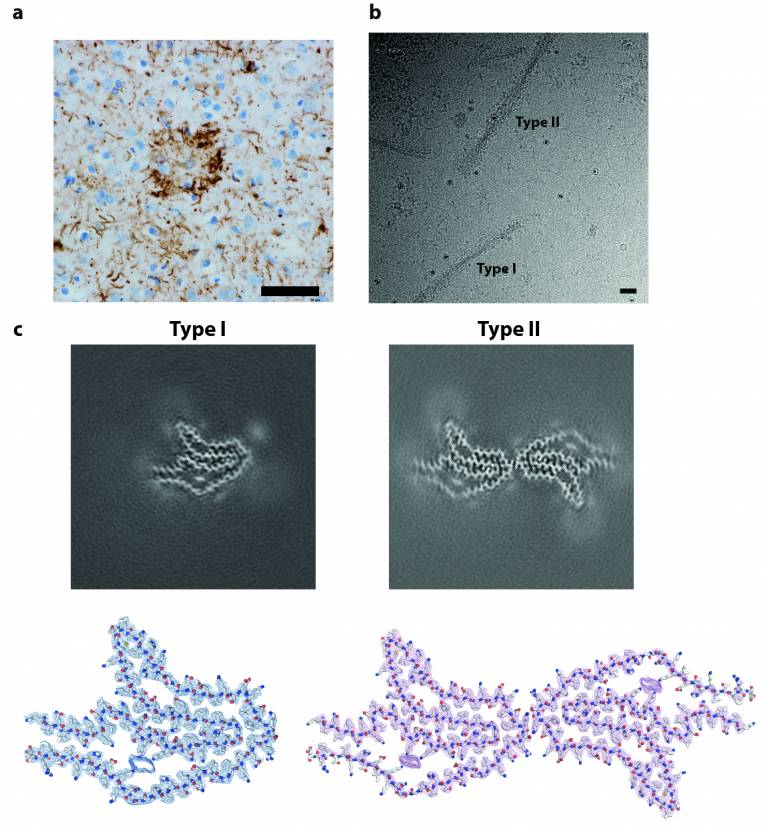
(a) Staining of neuronal inclusions, neuropil threads and astrocytic plaques in the frontal cortex of a patient with corticobasal degeneration (CBD) by antibody RD4 (specific for 4R tau, brown) (b) Cryo-EM image of tau filaments extracted from the frontal cortex of CBD patient (c) Cryo-EM maps of Type I and Type II tau filaments and their atomic models.
Selected Publications
Cryo-EM structures of amyloid-β 42 filaments from human brains.
Yang Y*, Arseni D*, Zhang W*, Huang M, Lövestam S, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Ryskeldi-Falcon B, Scheres SHW and Goedert M. Science. 2022;375(6577):167-172.
Structure-based Classification of Tauopathies
Shi Y.*, Zhang W. *, Yang Y., Murzin A.G., Falcon B., Kotecha A., Beers M., Tarutani A., Kametani F., Garringer H. J., Vidal R., Hallinan G.I., Lashley T., Saito Y., Murayama S., Yoshida M., Tanaka H., Kakita A., Ikeuchi T., Robinson A.C., Mann D.M.A., Kovacs G.G., Revesz T., Ghetti B., Hasegawa M., Goedert M., Scheres S.H.W. (2021) Nature 598(7880): 359–363 (*Contributed equally)
Novel tau filament fold in corticobasal degeneration
Zhang W., Tarutani A., Newell K.L., Murzin A.G., Matsubara T., Falcon B., Vidal R., Garringer H.J., Shi Y., Ikeuchi T., Murayama S., Ghetti B., Hasegawa M., Goedert M., Scheres S.H.W. (2020) Nature 580(7802): 283–287.
Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases
Zhang W., Falcon B., Murzin A.G., Fan J., Crowther R.A., Goedert M., Scheres S.H. (2019) . Elife. 8:e 43584
Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold
Falcon B. *, Zhang W. *, Schweighauser M., Murzin A.G., Vidal R., Garringer H.J., Ghetti B., Scheres S.H.W., Goedert M. (2018) Acta Neuropathol. 136(5): 699-708. (*Contributed equally)
Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10
Zhang W., Lukoyanova N., Miah S., Lucas J., Vaughan C.K. (2018) Cell Rep. 24(3): 744-754.
Crystal structure of the middle domain of SSRP1 reveals a role in DNA binding
Zhang W., Zeng F., Liu Y., Shao C., Li S., Lv H., Shi Y., Niu L., Teng M., Li X. (2015) Sci Rep. 5:18688.
Crystal structures and RNA-binding properties of the RRMs of hnRNP L: Insight into its roles in alternative-splicing regulation
Zhang W., Zeng F., Liu Y., Zhao Y., Lv H., Niu L., Teng M., Li X. (2013) J. Biol. Chem. 288(31): 22636-49.
 Close
Close

