Szymon W. Manka
 s.manka@ucl.ac.uk • PubMed • Google Scholar • |
Research Synopsis
We aim to define the long-sought prion pathogenicity mechanisms by single molecule localisation microscopy (SMLM), correlative light and cryogenic electron microscopy/tomography (cryo-CLEM/ET) and single particle electron cryo-microscopy (cryo-EM). This involves precise spatiotemporal monitoring of prion spread in live cells and identification of extra- and intra-cellular prion pathogenicity mediators by tracking fluorescently-labelled prions and their interaction partners.
The goal is to assign the roles of prion interaction partners at defined stages of prion infection, which may potentiate rational design of novel anti-prion therapeutics. Crucially, cellular prion protein (PrPC), besides being the substrate for prion (PrPSc) propagation, is likely a toxic acceptor of its misfolded self (PrPSc and potentially other forms) and of the Aβ oligomers or fibrils (the hallmark of Alzheimer's disease). This research might, therefore, have an immediate additional translational application in AD, with ensuing economic benefits.
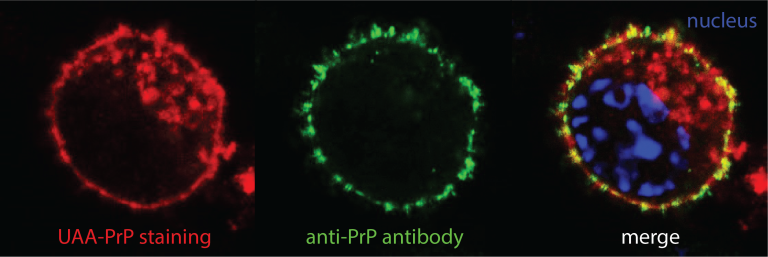
Genetic code expansion and bio-orthogonal labelling
We equip prion-propagating cell lines with a genetic code expansion (GCE) machinery for site-specific incorporation of an unnatural amino acid (UAA) into PrP. The UAA can be tagged with fluorescent dye suitable for super-resolution microscopy and cryo-CLEM/ET. Our recently solved cryo-EM structures of authentic infectious ex vivo RML and ME7 prion fibrils revealed a good candidate region for the UAA insertion.
The transgenic GCE cell platform will pave the way to the generation of a conditional GCE mouse expressing UAA-PrP. This will open a major opportunity to study fluorescent prion assemblies in situ by analysing mouse brain slices by cryo-CLEM/ET.
Selected Publications
Prion strains viewed through the lens of cryo-EM.
Cell Tissue Res. 2022 Aug 27. doi: 10.1007/s00441-022-03676-z.
Prion Propagation is Dependent on Key Amino Acids in Charge Cluster 2 within the Prion Protein.
A structural basis for prion strain diversity.
2.7 Å cryo-EM structure of ex vivo RML prion fibrils.
Pseudo-repeats in doublecortin make distinct mechanistic contributions to microtubule regulation.
A microtubule RELION-based pipeline for cryo-EM image processing.
Microtubule structure by cryo-EM: snapshots of dynamic instability.
The role of tubulin-tubulin lattice contacts in the mechanism of microtubule dynamic instability.
 Close
Close

