Covid-19 Drug Discovery projects
The new SARS-CoV-2 coronavirus that causes Covid-19 has led to a global pandemic with severe health and socio-economic consequences.
7 September 2020
Few countries in the world have been protected from the devastating effects of this new SARS virus strain, which are particularly severe in the elderly and those with pre-existing medical conditions.
Vaccines against Covid-19 are now available to more and more people worldwide but repurposing efforts for existing antiviral drugs was less successful. Therefore, an important strategy is to identify and develop new drugs that target SARS-CoV-2, to combat and contain current and future outbreaks.
We have established a network of scientists at the School of Pharmacy contributing to the understanding of SARS CoV-2 structure and function covering all steps of the early-stage drug discovery and development process. We are currently working on a range of non-structural proteins present in the SARs-CoV-2 genome. All three groups work closely together in this highly interdisciplinary academic drug discovery project.
What we are working on
Prof. Shozeb Haider uses computational modelling techniques including molecular dynamics simulations, enhances sampling and deep learning methods to contribute to the understanding the dynamics of SARS-CoV-2 proteins and complexes, the identification of inhibitor binding pockets, and the molecular docking of inhibitors to guide the synthesis of new inhibitor analogues to target Covid-19. For example, providing a dynamic profile for all known inhibitor-binding sites on the nsp15 (Figure 1) to develop pan-β-coronavirus main protease inhibitors. Other projects include computational modelling of a potential binding pocket at the interface between non-structural proteins 10 and 16/14. A current PhD student, Huan Wang is working on modelling Covid-19 proteins.
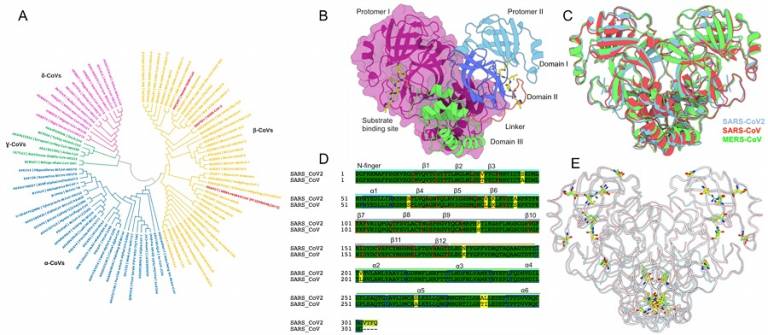
Figure 1: Overview of ß-coronavirus 3CL Mpro. (A) A phylogenetic tree of the α (blue), ß (yellow), γ (green) and δ (pink) coronavirus family; (B) Structure of the dimeric SARS-CoV2 Mpro enzyme (PDB 6LU7). The two protomers are represented in two different colors; the structural domains in protomer II are illustrated as cartoons; (C) Comparison of SARS-CoV2 (PDB 6LU7, cyan), SARS-CoV (PDB 2C3S, red) and MERS-CoV (PDB 4YLU, green) crystal structures; (D) Sequence alignment between SARS-CoV2 and SARS-CoV highlighting the position of 12 dissimilar residues in yellow. The structural elements have been annotated on the sequence and (E) Spatial position of the dissimilar residues (yellow, SARS-CoV2; green, SARS-CoV) highlighted on the Mpro structure.
Prof. Frank Kozielski’s group is interested in the early stages of drug development including inhibitor/fragment screening, the biophysical and biochemical characterisation of protein-inhibitor/fragment complexes, drug optimisation using structure-based drug design, and the determination of crystal structures of potential Covid-19 protein targets. Recently, we determined the crystal structure of SARS-CoV-2 nsp10 [2]. This structure (Figure 1) served as a basis for fragment-based screening [3]. Five PhD students, Danni Dong, Qian Wang, Xinyue Shi, Shumeng Ma and Jiaqi Lou, are working on various Covid-19 protein targets.
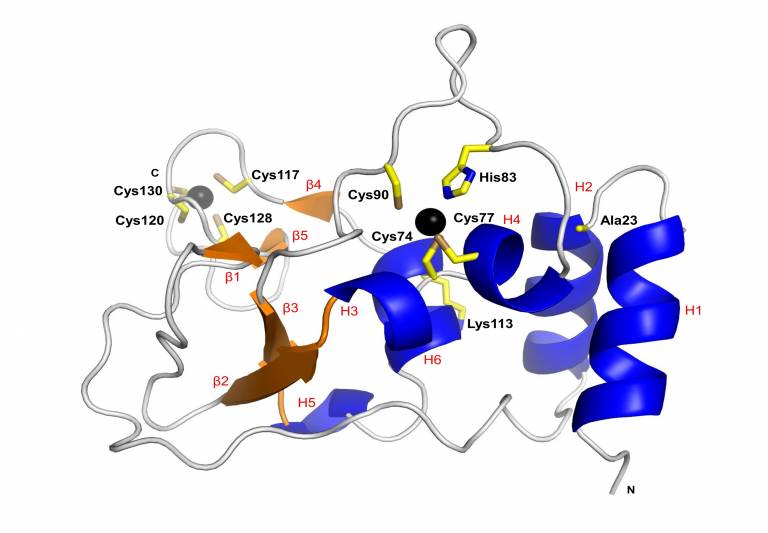
Figure 1: The crystal structure of unbound SARS CoV-2 non-structural protein 10 at 1.6 Å resolution. The structure will be used for structure-based drug design (Figure taken from [2].
[2] Rogstam A., Nyblom M., Christensen S., Sele C., Talibov V.O., Lindvall T., Rasmussen A.A., André I., Fisher Z., Knecht W., Kozielski F. (2020). Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2. Int. J. Mol. Sci.
[3] Kozielski F., Sele C., Talibov V.O., Lou J., Dong D., Wang Q., Shi X., Nyblom M., Rogstam A., Krojer T., Fisher Z. and Knecht W. (2021). Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16. RSC Chemical Biology, 2021.
We have several collaboration partners with whom we work together on various aspects of the larger project including Dr. Wolfgang Knecht (Lund University, Sweden); Dr. Zoe Fisher (European Spallation Source (ESS), Sweden); and Dr. Michela Mazzon (Virology Research Services Ltd).
Dr. Geoff Wells uses molecular modelling and medicinal chemistry approaches to design potential inhibitors of SARS-CoV-2 proteins. In addition to collaborating with Frank Kozielski and Shozeb Haider on projects relating to non-structural proteins (nsp) 10, 14 and 16, we have also worked in a consortium to target nsp13, the viral helicase protein that plays roles in RNA unwinding, proof reading and capping [4]. We have built molecular models of the RNA and/or ATP bound forms of the protein, both alone and in complex with other nsps. We have identified potential ligand binding sites in the ATP and RNA binding grooves and at allosteric/cryptic pockets on the protein surface and characterised the dynamics of these sites [5]. Our goal is to develop small molecule helicase inhibitors to act as antiviral agents in future outbreaks of Covid-19 and new emergent coronaviruses.
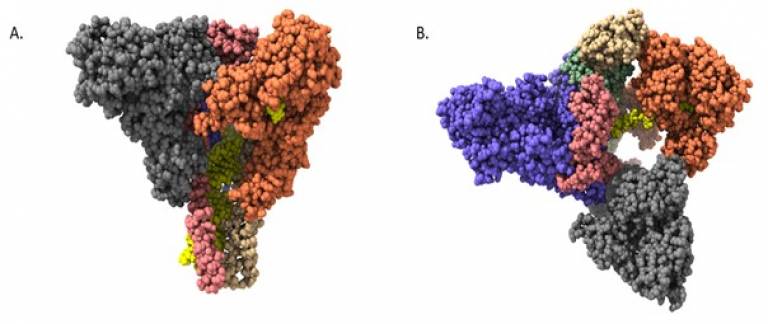
Figure 1. (A) Side view of the SARS-CoV-2 mini-replication-transcription complex composed of nsp 7, 8, 12 and two nsp13 units (the nsp13 helicase monomers are shown in orange and grey; the RNA is shown in yellow) and (B) top view (PDB ID: 7cxm).
[4] https://www.hecbiosim.ac.uk/covid-19-alliance
[5] Dénes Berta, Magd Badaoui, Pedro J. Buigues, Sam Alexander Martino, Andrei V. Pisliakov, Nadia Elghobashi-Meinhardt, Geoff Wells, Sarah A. Harris, Elisa Frezza, Edina Rosta. Modelling the active SARS-CoV-2 helicase complex as a basis for structure-based inhibitor design. Chem. Sci. 2021.
We are actively working on SARS-CoV-2 nsp1, nsp5, nsp10, nsp14 and nsp16. For enquiries about PhD projects or collaboration opportunities, please contact members of the Covid-19 research network.
 Close
Close

