The 3D structure of a protein can provide a sensitive probe of distant evolutionary relationships. Being able to identify similarities between protein structures can be a valuable tool in understanding aspects of evolution. A number of algorithms have been developed within the group to automatically assess the structural similarity between two proteins.
SSAP
The SSAP method (Taylor & Orengo, 1989) employs dynamic programming (DP) at two levels to cope with the possibility of extensive insertions and deletions between distantly related proteins. The first level of DP is employed for the comparison of residue structural environments between
proteins. The second level of DP is used in a final pass to determine the set of equivalent residues.
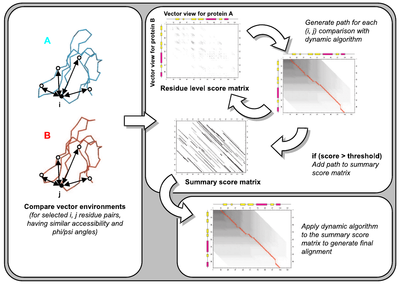
Figure used in textbook: Bioinformatics: Genes, Proteins and Computers (Orengo et al) (available on Amazon)
GRATH
GRATH employs graph theoretical techniques to identify equivalent secondary structure elements, i.e. those possessing similar intramolecular relationships. First, the 3D information embodied in a protein structure is reduced to a simplified 2D map, or graph, then well established graph theory techniques are employed to compare these graphs.
The simplified nature of the data makes this algorithm less sensitive than SSAP, but extremely fast.
CATHEDRAL
CATHEDRAL aims to combine the speed of GRATH and the accuracy of SSAP. Essentially GRATH is used to filter the full library of structures down to a small list that are potential matches. SSAP is then employed on this short list of matches to provide an accurate alignment and comparison score.
 Close
Close

