Drug-resistant gene goes from pig farms to patients worldwide
26 March 2018
A troublesome gene that is resistant to an antibiotic often used as a last resort has been tracked from its origins to hospital patients worldwide in a new study led by UCL and Peking University People's Hospital.
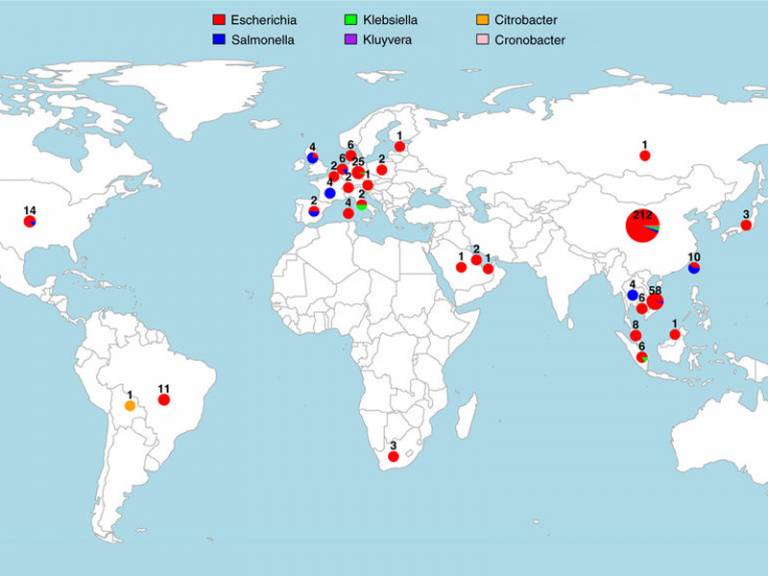
The study, published in Nature Communications, found that the mcr-1 gene, now present across the globe, can be tracked to a single event around 2006 when it moved from pigs into pathogens that affect humans.
"The speed at which mcr-1 spread globally is indeed shocking," said the study's lead author, Professor Francois Balloux (UCL Genetics, Evolution & Environment).
The mcr-1 gene makes bacteria resistant to colistin, which is one of the very few drugs effective against multi-drug resistant infections. Colistin was discovered in the 1950s but has until recently been mostly used in pig farming due to its severe side-effects. With the recent increase in antibiotic resistance, it has now become widely prescribed in the clinic as a last-line drug for infections such as E. coli and Klebsiella pneumoniae.
"Mcr-1 provides relatively low resistance, and mcr-1 strains can still be treated with colistin, but at far higher doses, which is obviously detrimental to the patients as colistin is fairly toxic. I wouldn't be surprised if we saw the emergence and spread of elements that provide higher resistance to colistin than mcr-1, which would make the situation even worse," said Professor Balloux.
The mcr-1 gene is mobile and plasmid-borne, meaning it can be transported from one bacterium to another, even of two different species, and confer resistance to colistin. It was identified in China in 2016, followed by a rapid realisation that it had already spread globally, but its exact origin and spread had not yet been determined.
The research team compiled an exhaustive dataset of 457 mcr-1 positive genome sequences isolated from humans and farm animals from five continents, by sequencing the genomes of 110 bacterial strains and systematically mining previously deposited genomic data from publicly available databases.
They analysed this data with novel computational tools to show there had been a single emergence of mcr-1, reliably dated to the mid-2000s. They also reconstructed how the mcr-1 element had been spreading globally and to various bacterial pathogens by hitchhiking with various bacterial mobile genetic elements.
This work represents the first reconstruction of the emergence and spread of an antibiotic resistance element and opens new avenues for improved global surveillance of antibiotic resistance using genomic tools.
Hospitals worldwide are struggling with increasing incidence of hard-to-treat antibiotic-resistant bacterial infections. The World Health Organisation has been raising the alarm over a post-antibiotic era where minor infections could routinely become untreatable. Public Health England predicts that antibiotic resistance could lead to 10 million deaths every year globally by 2050 and to £66 trillion in lost productivity to the global economy.
"There is no consensus yet on the importance of antimicrobials in livestock being a main driver of the antimicrobial resistance public health crisis. That said, the use of antimicrobials in agriculture is not only a scientific issue but also a societal and economic issue that will need to be addressed," said Professor Balloux.
"Given the dearth of new antibiotics in the pipeline, our best hope to avert the current public health crisis is to improve stewardship of existing drugs, by harnessing the potential of bacterial genome sequencing and translate it into improved surveillance and diagnostics tools," he added.
"Our study highlights the value of analysing DNA sequenced from hundreds of resistant bacteria to track the spread of dangerous resistance genes around the world. By deciphering the genetic code of these bacteria we found it was possible to predict not only how and where but also when mcr-1 started to spread. This is so important as the presence of mcr-1 across the globe, in many different bacteria species, all within only a decade highlights the ease and speed with which these resistant genes can be disseminated," said co-author Dr Lucy van Dorp (UCL Genetics, Evolution & Environment).
"Our work suggests that mcr-1 was already distributed worldwide just a decade after its emergence on a mobile genetic element. While we don't know the exact transmission routes it spread by, we can presume that other AMR genes will be spread in a similar way, emphasizing that AMR is a truly global problem requiring coordinated international action," added co-author Liam Shaw (UCL Genetics, Evolution & Environment).
The study team involved researchers from UCL, Peking University People's Hospital, University of Oxford, Shandong Academy of Agricultural Sciences, CIRAD, University of Edinburgh, University of Cambridge and Imperial College London. The research was funded by the Newton Trust, NSFC, BBSRC, EPSRC, Wellcome, the European Union and the Royal Society.
Links
- Research paper in Nature Communications
- Professor Francois Balloux's academic profile
- UCL Genetics, Evolution & Environment
- UCL Life Sciences
- Media coverage
Image
- Global distribution of mcr-1. Source: R Wang et al.
Media contact
Chris Lane
Tel: +44 (0)20 7679 9222
Email: chris.lane [at] ucl.ac.uk
 Close
Close

