New antibiotics a step closer with discovery of bacterial protein structure
2 June 2011
Scientists have uncovered the structure of the protein complex that assembles the tiny hair-like strands that cover the outside of bacteria.
 Called pili, these 'hairs' allow bacteria to
group together and stick to human cells to cause infection - and are therefore
a key target for a new generation of antibiotics.
Called pili, these 'hairs' allow bacteria to
group together and stick to human cells to cause infection - and are therefore
a key target for a new generation of antibiotics.
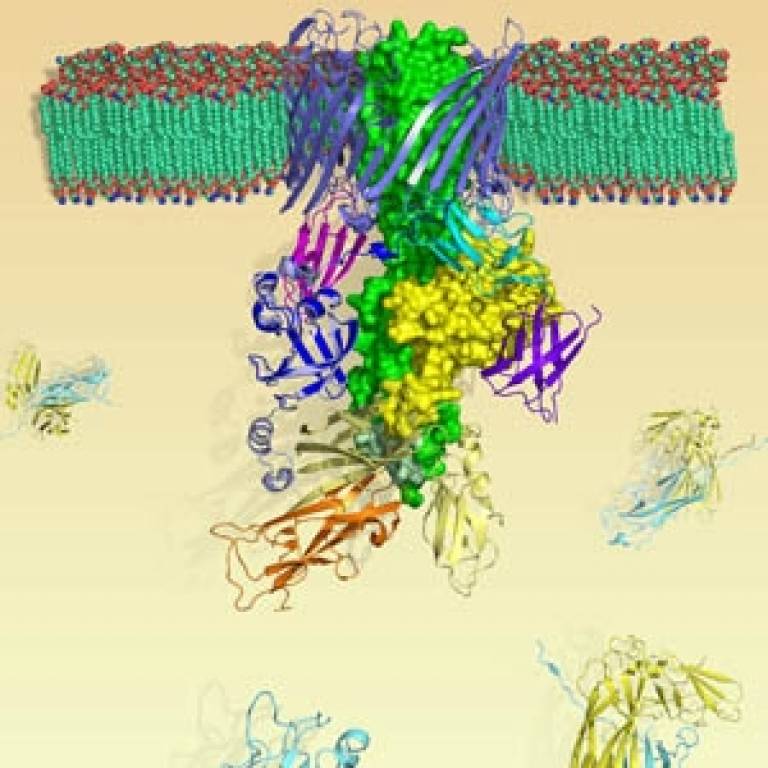
Published in Nature, scientists at the Institute of Structural and Molecular Biology (a joint institute between University College London (UCL) and Birkbeck) have revealed the structure of a complex protein called FimD that acts as an assembly platform for the pili of cystitis bacteria. The structure of the FimD protein means scientists can see the process of pili assembly, from individual protein subunits to complete structures, for the first time.
Pili are tiny hair-like strands on the outside of bacteria that help them to link together in groups. In the case of cystitis, pili allow bacteria to attach themselves to the wall of the bladder, leading to bladder cells engulfing the bacterium. Once the bacteria have invaded the bladder cells, they escape traditional antibiotic treatment and lie dormant, making recurrent infections very common.
Scientists believe that antibiotics could be developed that disrupt the FimD protein, and therefore the production line of pili proteins. The UCL/Birkbeck group, together with collaborators in the USA, have already discovered small molecules able to interfere with pilus biogenesis. Without their pili, bacteria cannot attach to each other or the host, making infection much less likely.
Professor Gabriel Waksman from the UCL Research Department of Structural and Molecular Biology and the Birkbeck Department of Biological Sciences, who led the research, said: "Cystitis is one of the most common gram negative bacterial infections; it can also be extremely painful and surprisingly hard to treat - especially repeat infections."
"Pili are a prime target for a new breed of antibiotics to target cystitis and other conditions, as without pili bacteria are unable to attach themselves to each other or the walls of human cells, and therefore much less likely to cause serious infections."
Pili protein subunits are made inside bacteria and initially transported through the inner cell wall. Each subunit is then picked up by a 'chaperone' protein which takes it across to a protein in the outer cell wall called the 'usher'.
The usher protein coordinates the ordered assembly of pilus subunits, i.e. the growth of the pili. This research, funded by the Medical Research Council, has isolated and crystallised the usher protein in cystitis bacteria, FimD, while it's bound to the chaperone/subunit combination.
The structure of FimD provides insights into pilus biogenesis because it unravels the entire mechanism of subunit polymerization and transport across the outer wall of the bacteria. "Scientists have been working for a number of years to work out the how pili are assembled. This is the first view of the transportation and assembly of pili in action," said Professor Waksman.
Media contact: Clare Ryan
Image: Structure of the FimD protein complex
UCL context
The UCL Research Department of Structural and Molecular Biology aims to provide a stimulating and diverse research and
training environment of international standing within which important
and exciting areas of modern biology, biotechnology and medicine can be
investigated at the atomic, molecular, cellular and organism level.
The department pursues these research aims through its high level of external research funding, its investment in excellent new facilities, up-to-date equipment and state-of-the-art technologies, the recruitment of high-calibre staff, maintenance of industrial contacts, and the fostering of close links and collaborations within the extensive UCL and Bloomsbury scientific community.
 Close
Close

