New structure of Alzheimer's drug predicted by e-Materials project
10 January 2006
The UCL-led e-Materials project has achieved one of the holy grails of the pharmaceutical industry - the computational prediction of a previously unidentified crystal structure, or polymorph, of a drug molecule.
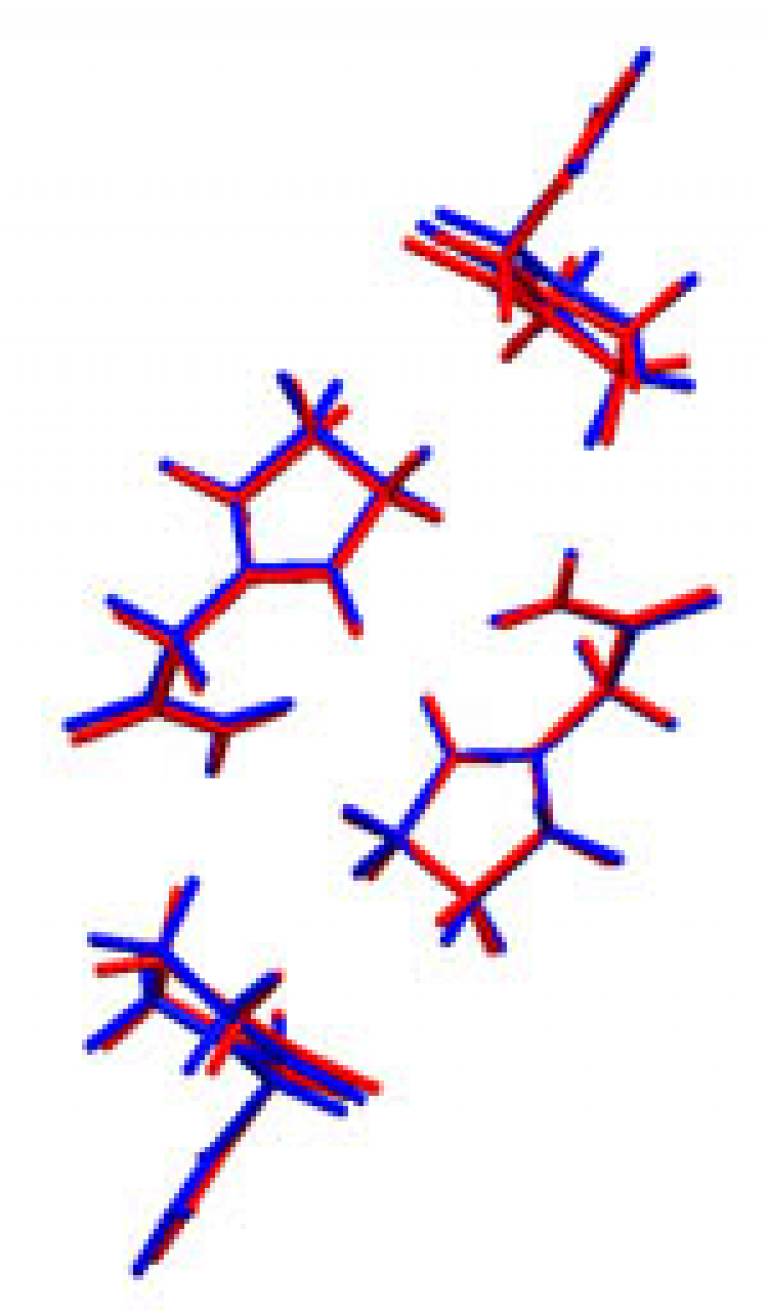
Researchers working on the project have successfully predicted a new polymorph of the Alzheimer's drug, Piracetam. The action of a drug is dependent not only on its chemical composition, but also on the way in which the drug molecules arrange themselves. For example, crystal structure can affect the drug's solubility and hence its rate of absorption into the bloodstream. An unexpected polymorph could alter the drug's therapeutic properties if it inadvertently contaminated the standard formulation.
Polymorphs are the bane of the pharmaceutical industry because they can be difficult to predict. Industrial chemists put a lot of effort into searching for new polymorphs using experimental techniques, but they can never be sure they have found them all. A new polymorph can turn up years later, sometimes resulting in the withdrawal of a drug from the market whilst the problem is identified and solved. Pharmaceutical companies are also keen to patent every polymorph of a drug to prevent a rival from undercutting them later with a new, therapeutically effective version.
The e-Materials project, funded by the Engineering and Physical Sciences Research Council (EPSRC) as part of the e-Science Core Programme, applied Grid technologies to address this problem. "We chose to study Piracetam because it's a well understood drug with three known polymorphs. We thought it was a good one to test our new methods against," said Professor Sally Price (UCL Chemistry).
After their work was presented at the 2004 e-Science All Hands meeting, Professor Price was presented with a challenge. Dr Colin Pulham (University of Edinburgh) asked her to predict the crystal structure of a new polymorph of Piracetam that he had discovered using high-pressure crystallisation techniques. He said: "I told Professor Price that we had discovered a new form, but I didn't tell her what it was. If her techniques were effective, she should have been able to find it."
Predicting polymorphism of organic crystal structures is computationally very demanding. Millions of possible structures need to be analysed to identify those that are likely to be the most stable. Dr Harriott Nowell (UCL Chemistry) and Professor Price conducted the piracetam analysis on the UCL Research Computing C^3 cluster, using a workflow developed by Mr Ben Butchart and Professor Wolfgang Emmerich (UCL Computer Science). A few months after her challenge, Professor Price submitted a list of candidate structures in order of probability. The first on the list matched the structure that Dr Pulham and his team had found experimentally. He said: "It was bang, spot on." Professor Price added: "This result does a lot for the credibility of our methodology."
The e-materials project is building up a database containing the outputs of computational polymorphism searches at the Central Laboratory for the Research Councils (CCLRC). The database and a data portal will shortly be available on the National Grid Service, providing information on an increasing range of molecules that may help identify other new polymorphs. Indeed, a new polymorph of aspirin has recently been discovered that corresponds to one of the structures discovered in the earliest study of this project.
The project is continuing to increase the range of molecules whose possible polymorphs are being computed by Professor Emmerich's group, developing the use of the Business Process Execution Language (BPEL) for Condor environments, such as the UCL Research Computing Condor Pool. The computed results are being compared with experimental screens for new polymorphs at UCL Chemistry and University of Strathclyde Pharmaceutical Sciences, with Dr Pulham sometimes collaborating with his high pressure experiments. Thus the computational work at UCL is underpinning a major project on polymorphism, Control and Prediction of the Organic Solid State (CPOSS), funded by the Basic Technology Program of the Research Councils UK.
Image: Illustration of how closely matched the experimental (red) and predicted (blue) polymorph structures are.
 Close
Close

