Methodological dangers of a dental calculus microbiome analysis
12 November 2018
Since at least the 1980s, it has been known that archaeological dental calculus contains preserved cellular structures of oral bacteria, but it was only recently discovered that it is also a robust and long-term reservoir of well-preserved DNA (Adler et al.
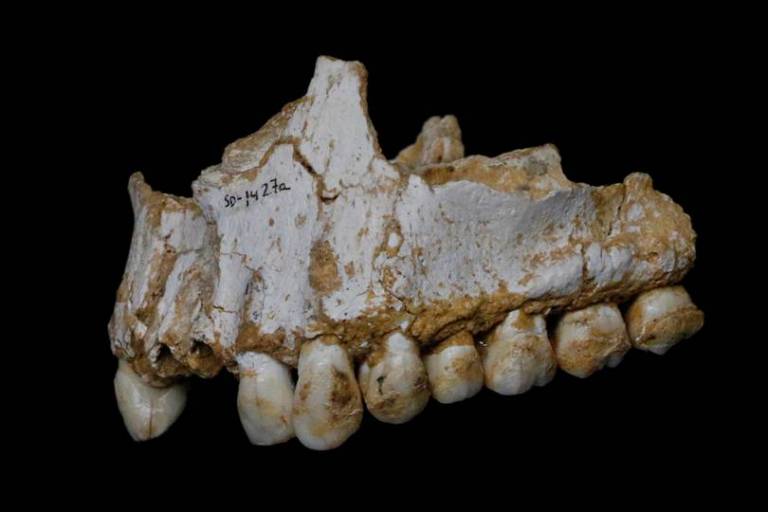 , 2013). Advances in ancient DNA now enable direct comparisons between ancient and modern oral microbial communities. Recently, Weyrich et al. (2017) suggested that preserved dental calculus could be a useful source of information for the reconstruction of Neanderthal behavior, diet, or disease. The authors succeeded in deeply sequencing five Neanderthal individual dental calculus samples, retrieving in three of them (one individual did not provide any genetic data, another was omitted because of possible contamination with modern humans) 93.76% of bacterial sequences, 5.91% archaeal sequences, 0.27% eukaryotic sequences, and 0.06% viral sequences. Shotgun-sequencing of ancient DNA from these specimens brought to light regional differences in Neanderthal ecology: For instance, at Spy Cave, Belgium, a heavily meat-based diet (including woolly rhinoceros and mouflon) was evident, which is characteristic of a steppe environment, whereas at El Sidrón Cave, Spain, no meat eating was detected, but mushrooms, pine nuts, and moss were eaten, reflecting forest gathering. Weyrich et al. (2017) suggested that differences in diet were linked to an overall shift in the oral microbiota, and proposed that meat consumption may have contributed to substantial variation within Neanderthal microbiota. From these dental microbiome data, and notably from phylogenetic analyses of an Archaea species named Methanobrevibacter oralis (10.2 × depth of coverage, the oldest draft microbial genome generated to date, at ∼48 ka), the authors inferred interbreeding between modern humans and Neanderthals. Based on the molecular clock and a comparison between M. oralis subsp. neandertalis, isolated from a Neanderthal genome, and M. oralis, isolated from modern humans, they concluded that the divergence of these microbial subspecies occurred 143-112 ka, i.e., much later than the divergence between Homo sapiens and H. neanderthalensis (450-700 ka; Stringer, 2016). Based on these dates, Weyrich et al. (2017) concluded that these microorganisms could have been transferred between these hominins during interactions subsequent to their divergence, leading to the inference that modern humans and Neanderthals interbred.
, 2013). Advances in ancient DNA now enable direct comparisons between ancient and modern oral microbial communities. Recently, Weyrich et al. (2017) suggested that preserved dental calculus could be a useful source of information for the reconstruction of Neanderthal behavior, diet, or disease. The authors succeeded in deeply sequencing five Neanderthal individual dental calculus samples, retrieving in three of them (one individual did not provide any genetic data, another was omitted because of possible contamination with modern humans) 93.76% of bacterial sequences, 5.91% archaeal sequences, 0.27% eukaryotic sequences, and 0.06% viral sequences. Shotgun-sequencing of ancient DNA from these specimens brought to light regional differences in Neanderthal ecology: For instance, at Spy Cave, Belgium, a heavily meat-based diet (including woolly rhinoceros and mouflon) was evident, which is characteristic of a steppe environment, whereas at El Sidrón Cave, Spain, no meat eating was detected, but mushrooms, pine nuts, and moss were eaten, reflecting forest gathering. Weyrich et al. (2017) suggested that differences in diet were linked to an overall shift in the oral microbiota, and proposed that meat consumption may have contributed to substantial variation within Neanderthal microbiota. From these dental microbiome data, and notably from phylogenetic analyses of an Archaea species named Methanobrevibacter oralis (10.2 × depth of coverage, the oldest draft microbial genome generated to date, at ∼48 ka), the authors inferred interbreeding between modern humans and Neanderthals. Based on the molecular clock and a comparison between M. oralis subsp. neandertalis, isolated from a Neanderthal genome, and M. oralis, isolated from modern humans, they concluded that the divergence of these microbial subspecies occurred 143-112 ka, i.e., much later than the divergence between Homo sapiens and H. neanderthalensis (450-700 ka; Stringer, 2016). Based on these dates, Weyrich et al. (2017) concluded that these microorganisms could have been transferred between these hominins during interactions subsequent to their divergence, leading to the inference that modern humans and Neanderthals interbred.
Interbreeding between Neanderthals and modern humans: Remarks and methodological dangers of a dental calculus microbiome analysis
Philippe Charlier, Frédérick Gaultier, Geneviève Héry-Arnaud
DOI: 10.1016/j.jhevol.2018.06.007
 Close
Close

