Supervisors: Professor Waseem Qasim, Dr Christos Georgiardis
Background:
CD40 ligand (also known as CD40L or CD154) is a transmembrane protein of the TNF superfamily expressed primarily by activate T cells during inflammatory responses [1]. It interacts with CD40 which is a transmembrane co-stimulatory receptor expressed on B cells, dendritic cells, monocytes, and other non-haematopoietic cells. This interaction is vital for the development and function of B cells, effective immunoglobulin isotype switching for generation of long-lived plasma cells [2].
CD40L deficiency is a rare but life threatening x-linked immunodeficiency by more than 150 unique mutations within the CD40 ligand gene (CD40LG) [1,3]. This condition is characterised by opportunistic infections such as Pneumocystis pneumonia and Cryptosporidium colonisation. Children suffering from this x-linked hyper-IgM syndrome have a defective immune system as a result of the mutations in their CD40LG from the surface of their T cells. Conventional retroviral gene therapy failed at the modelling stage for CD40L with leukaemia arising from unregulated expression [2].
Gene editing techniques for CD40L correction have been reported using transcription activator-like effector nuclease (TALEN) reagents with successful restoration of a therapeutic DNA template [4]. Similar approaches are being developed with CRISPR/Cas9 [5].
Hypothesis:
Emerging technology such as CRISPR/Cas9 in the field of gene editing may be used to correct the mutated CD40L gene with the T cells of the affected people. The idea is to utilise this technology replace mutated CD40L exons with normal CD40L sequence. Emerging base editing and prime editing applications will be investigated.
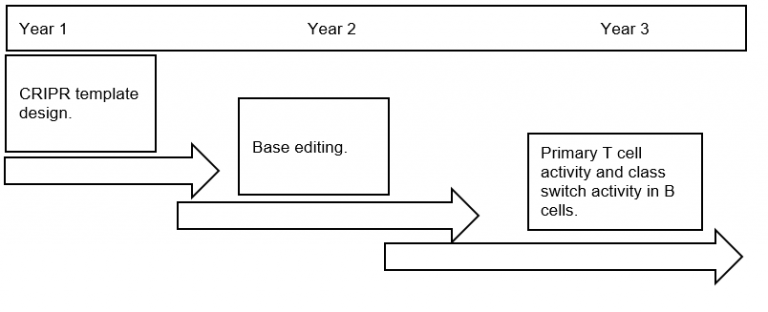
Timeline/Plan:
Year 1:
Generation and testing of CRISPR/Cas9 reagents:
Initial work will focus on designing and validating CRISPR synthetic RNAs (sgRNAs) with proximity to patients with mutations causing CD40L deficiency. This work will be done in T cell lines and primary T cells from healthy donors where CRISPR/Cas9 reagents will be delivered as RNA complex by electroporation. Targeted breakage at the CD40LG locus will be assessed at the genomic level through established Sanger sequencing and next generation sequencing (NGS) methods. Disruption of CD40L surface expression will be measure by flow cytometry following stimulation of cells with PMA and ionomycin.
Design and delivery of therapeutic template:
The challenging aspect of the approach we intend to use involves designing a short portion of DNA encoding specific exon 5 in the CD40L sequence, modified to prevent its cleavage by the CRISPR gRNAs, which will act as a template for the site specific correction of these mutations. Co-delivery of template DNA alongside the CRISPR/Cas9 machinery will instead trigger repair via the homology-directed repair (HDR) pathway thereby replacing mutated nucleotides with wild-type sequence. This approach could be applied to the majority of CD40L deficient patients.
Year 2:
Targeted correction of individual mutations:
Base editing technology allows for the chemical deamination of individual nucleotides within a restricted activity window [4]. Through the use of cytidine or adenosine deaminases C to T and A to G conversions or can be achieved opening the possibility of seamless correction without the associated risks of dsDNA breaks. Bespoke sgRNA sequences will be designed for each patient point mutation tailored to the appropriate base editor to allow reversion to the WT nucleotide or restoration of amino acid sequence. Where a conventional NGG protospacer associated motif (PAM) is not present in proximity to the mutation, evolved Cas9 variants with broader PAM recognition will be utilised. Therapeutic delivery for exon 5 will be performed using single-stranded oligo donor as they are well established and exhibit low toxicity upon their delivery.
Year 3:
Correction of primary T cells:
Optimised reagents from years 1-2 will be applied to patient specific mutations. Assays to confirm class switching activity will be established. The ability to scale and test the approach will be investigated.
Ethics Approval:
Ethical approval for collection of human samples for PID is in place at GOS and will be modified if required.
References:
1. Franca TT, Barreiros LA, Al-Ramadi BK et al (2019). CD40 ligand deficiency: treatment strategies and novel therapeutic perspectives. Expert Review of Clinical Immunology. 15(5):529-540.
2. Brown MP, Topham DJ, Sangster MY, et al. (1998). Thymic lymphoproliferative disease after successful correction of CD40 ligand deficiency by gene transfer in mice. Nat Med. 4(11): 1253-1260.
3. Hubbard, N., Hagin, D., Sommer, et al. (2016). Targeted gene editing restores regulated CD40L function in X-linked hyper-IgM syndrome. Blood, 127(21), 2513-2522.
4. Komor A.C., Kim Y.B., Packer M.S., et al. (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage.
5. Wang R, Graham S, Sun N, McCarthy D, et al (2020). CRISPR/Cas9-targeting of CD40 in hematopoietic stem cells limits immune activation mediated by anti-CD40. PubMed 10;15(3): e0228221.
 Close
Close




