The Proteomics Standard Initiative Common QUery InterfaCe (PSICQUIC) specification was created by the Human Proteome Organization Proteomics Standards Initiative (HUPO-PSI) in 2008. Its web service provides computational access to molecular-interaction data resources. The PSICQUIC protein interaction datasets are accessible directly from the Cytoscape molecular interaction network tool, allowing users to select the interaction data from all or a subset of the datasets available.
Recommendations for use When extracting protein interaction data, for example when using Cytoscape, we recommend that users consider including IntAct, BHF-UCL, EBI-GOA-nonIntAct and EBI-GOA-miRNA datasets.
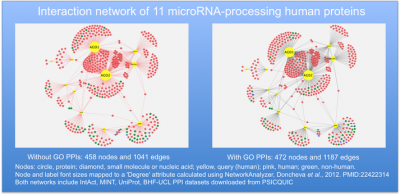
Describing the methods used to create a protein interaction network
It is important to consider that all scientific data should be reproducible. This is very difficult to do when the databases used in an analysis are updated on a regular basis. We recommend that the following information is included in methods sections:
- the tool used to create the network (e.g. Cytoscape) with a published citation (e.g. Su et al. 2014)
- the annotation dataset(s) included in the analysis (e.g. PSICQUIC interactions provided by IntAct, BHF-UCL, UniProt, EBI-GOA-nonIntAct, EBI-GOA-miRNA, MINT) with a reference if possible (e.g. Orchard et al. 2014)
- the date the analysis was performed. Note that some resources do provide dated datasets which would allow the analysis to be repeated by a third party
- information about any specific filtering that was applied during the analysis
- The identifiers (IDs) used in the analysis, mapping datasets IDs is not 100% accurate, providing the IDs will make it easier for a third party to repeat your analysis
References describing PSICQUIC and IntAct
- Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, Duesbury M, Dumousseau M, Galeota E, Hinz U, Iannuccelli M, Jagannathan S, Jimenez R, Khadake J, Lagreid A, Licata L, Lovering RC, Meldal B, Melidoni AN, Milagros M, Peluso D, Perfetto L, Porras P, Raghunath A, Ricard-Blum S, Roechert B, Stutz A, Tognolli M, van Roey K, Cesareni G, Hermjakob H. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014 Jan;42(Database issue):D358-63. PMID:24234451
- del-Toro N, Dumousseau M, Orchard S, Jimenez RC, Galeota E, Launay G, Goll J, Breuer K, Ono K, Salwinski L, Hermjakob H. A new reference implementation of the PSICQUIC web service. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W601-6. PMID:23671334
- Orchard S. Molecular interaction databases. Proteomics. 2012 May;12(10):1656-62. PMID:22611057
- Aranda B, Blankenburg H, Kerrien S, Brinkman FS, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, Gaulton A, Goll J, Hancock RE, Isserlin R, Jimenez RC, Kerssemakers J, Khadake J, Lynn DJ, Michaut M, O'Kelly G, Ono K, Orchard S, Prieto C, Razick S, Rigina O, Salwinski L, Simonovic M, Velankar S, Winter A, Wu G, Bader GD, Cesareni G, Donaldson IM, Eisenberg D, Kleywegt GJ, Overington J, Ricard-Blum S, Tyers M, Albrecht M, Hermjakob H. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat Methods. 2011 Jun 29;8(7):528-9. PMID:21716279
- Orchard S, Albar JP, Deutsch EW, Eisenacher M, Binz PA, Hermjakob H. Implementing data standards: a report on the HUPOPSI workshop September 2009,Toronto, Canada. Proteomics. 2010 May;10(10):1895-8. PMID:20623474
A collaboration between the UCL functional gene annotation team and the UniProt-GOA group at the EBI, funded by the British Heart Foundation, has led to the creation of a new protein interaction dataset, "EBI-GOA-nonIntAct" which can be accessed at PSICQUIC and included in network analyses.
The EBI-GOA-nonIntAct dataset includes all of the protein interaction data that has been created by the Gene Ontology (GO) Consortium, with all of the protein interaction data imported from IntAct removed.
In July 2019 the EBI-GOA-nonIntAct dataset included over 72,000 protein interactions, around 25% of which involve human proteins, although almost 200 distinct taxons have been annotated.
The Gene Ontology interaction data is created in a very simple format i.e. protein A binds protein B supported by the evidence code IPI (Inferred from Physical Interaction), as described in a specific publication. Whereas, the protein interactions created by IntAct apply the IMEx consortium standards and therefore provide very detailed information about each protein interaction, such as the constructs and methods used to identify the interaction. The GO Consortium annotation dataset includes annotations imported from IntAct as well as other databases.
Why was the IntAct data removed from the EBI-GOA-nonIntAct dataset?
The protein interaction data provided by IntAct is of a much higher and more descriptive standard than the GO annotation data. However, when the IntAct data is imported into the GO annotation dataset the interactions are all imported with the IPI evidence code. Network analysis tools, such as Cytoscape, are not able to identify duplicate annotations that are present in both IntAct and the GO annotation dataset. We have therefore decided to provide only the GO consortium created protein interaction annotations in the EBI-GOA-nonIntAct dataset. This then enables uses to combine the highly descriptive IntAct annotations with the more simple GO consortium data.
The BHF-UCL functional gene annotation curators have submitted over 4,000 IMEx standard protein interaction data using the IntAct editing tool. This data is also available on the PSICQUIC website.
A collaboration between the UCL functional gene annotation team and the UniProt-GOA group at the EBI, funded by the British Heart Foundation, has led to the creation of a new miRNA:mRNA association dataset, "EBI-GOA-miRNA" which can be accessed at PSICQUIC and included in network analyses. To associated different miRNA identifiers with the annotations in this file download mapping files from the RNAcentral FTP site: ftp://ftp.ebi.ac.uk/pub/databases/RNAcentral/current_release/id_mapping/database_mappings/. The tarbase file has the miR-##-## symbol format, whereas the hgnc file has the HGNC symbols.
MicroRNAs act by base pairing with an mRNA target and affecting the expression of the mRNA or translation of protein from the mRNA. Experimental assays can be used to test the effect of the miRNA on the mRNA, either by demonstrating direct binding between the two (as in the case of a reporter assay) or a change in level of the mRNA in response to the miRNA (e.g. a qRT-PCR assay). We curate these associations between the miRNA and the mRNA using the following GO terms, depending on the experimental evidence available in the paper:
Biological Process
GO:0035195 gene silencing by miRNA
GO:0035278 miRNA mediated inhibition of translation
GO:0035279 mRNA cleavage involved in gene silencing by miRNA
GO:0098806 deadenylation involved in gene silencing by miRNA
E.g. Human miR-30e-5p is involved in gene silencing by miRNA of human ADRB1.
Annotation: URS00001DE669_9606 GO:0035195 IDA PMID:25950484 has_input (P08588)
Molecular Function
GO:1903231 mRNA binding involved in posttranscriptional gene silencing (used only for reporter assays)
E.g. Human miR-30e-5p binds the human BNIP3L mRNA in order to silence it.
Annotation: URS00001DE669_9606 GO:1903231 IDA PMID:25950484 has_direct_input(ENSG00000104765)
In March 2022 there were 4,400 miRNA:mRNA associations in this dataset.
 Close
Close