Editor - Barbara Kramarz: I am pleased to take on the role of editor for our first newsletter introducing the GO Annotation projects funded by Alzheimer's Research UK
Current amyloid-ß-focused annotation project
Our current six-month project, funded by Alzheimer's Research UK (ARUK), focuses on Gene Ontology (GO) annotation of amyloid-ß (Aß) cell surface receptors and other proteins involved in clearance of Aß and mediating the neurotoxicity of Aß oligomers (AßO). The proteins to be annotated within the next six months are listed in Table 1 in Jarosz-Griffiths et al., 2016, JBC (PMID:26719327), or in Figure 1. The current grant is jointly held by Dr Ruth Lovering, an expert GO curator at UCL, and Professor Nigel Hooper, the Director of Dementia Research at the University of Manchester, and the senior author of the Jarosz-Griffiths et al. review. I will be creating the majority of the GO annotations, whereas Tony Sawford, a software engineer from the EMBL-EBI, ensures smooth running of the GO annotation tool.
|
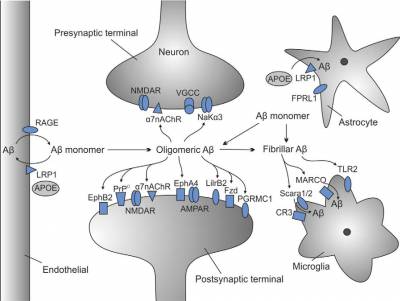 Figure 1. Aβ receptors and their cellular locations. (Figure from Jarosz-Griffiths et al., 2016, JBC). |
Future Alzheimer's
Research UK-funded work
We are pleased to announce that upon completion of the Aß-focused project, our team will continue to annotate Alzheimer's disease-relevant biology, thanks to a recently awarded three-year ARUK grant. The grant is jointly held by Dr Ruth Lovering (PI), Professor Nigel Hooper and Dr David Brough from the University of Manchester, Dr Rina Bandopadhyay (UCL), as well as Dr Maria-Jesus Martin and Dr Helen Parkinson from the EMBL-EBI. The goal of this three-year project will be to annotate gene products and processes associated with tau protein pathology and neuroinflammation.
Protein Complex data
In order to annotate the roles of Aß receptors and their full biological context using GO, curators need to be able to specify the types and/or conformations of Aß being annotated. Currently it is possible to capture information about the roles of specific monomeric Aß peptides only, using UniProt identifiers. For instance, human Aβ1-40 and Aβ1-42 monomers are represented using UniProt identifiers P05067:PRO_0000000093 and P05067:PRO_0000000092, respectively. However, in order to annotate AßO, the primary Aß species responsible for mediating neurotoxicity, new EBI Protein Complex identifiers first need to be generated. I have been liaising with neurobiology experts and the EBI Protein Complex curators to review experimental evidence describing structures and functions of the different types of AßO to generate the Protein Complex database records and identifiers, allowing for GO annotation of these oligomers and their receptors.
Gene annotation
Based on the EBI statistics, 17th March 2017, the ARUK-UCL gene annotation team has associated 268 GO annotations with 48 gene products, 162 of which are to 18 human gene products.
Meetings attended
In March Barbara attended the ARUK Research Conference 2017 in Aberdeen, thanks to a Travel Award from the ARUK Research Network. Barbara presented a poster, entitled Capturing amyloid-ß-relevant knowledge with Gene Ontology, which provided an overview of our current GO annotation project, its milestones, challenges and achievements to date.
12-13 June: Introduction to Bioinformatic Resources and GO workshop
Please register for this two-day workshop using the Eventbrite link. This free workshop, funded by the British Heart Foundation (BHF), provides an overview of the available databases and resources as well as the GO curation process. Our group will answer your questions and help with your GO data analysis.
Please contact us to receive this quarterly Newsletter by email or to ask any questions about our project.
 Close
Close








