Project Leader: Liselotte Backdahl
Inflammatory bowel disease (IBD), is characterized by a complex aetiology in a combination of genes and environment. Despite years of research very few disease causing factors have been identified. This extended complexity could be attributed both to disease pathway heterogeneity and to epigenetic modifications of the genomic landscape that leads to altered gene expression profile of the disease tissue. The epigenetic machinery consists of short interfering RNA, histone modifications and DNA methylation. New tools are now available that will enable studies of these phenomena. By combining immuno-precipitation of methylated Cytosine containing DNA with a high resolution tiling array, consisting of 390K 50mer probes of significant epigenetic importance, a comprehensive insight to the DNA methylation profile of an individual in a given cell type can be generated. The ideal way to investigate "non-genetic" disease risk-factors is to study monozygotic twins and epigenetic and transcriptional differences and the transcription/ methylation levels will be analysed in mucosa biopsies.
Epigenetic regulation of inflammatory bowel disease
Inflammation is a key biological mechanism in the organism's defence system against harmful intruders. However, dysregulation of inflammatory mechanisms may be detrimental and is the underlying cause for widespread chronic diseases such as asthma, psoriasis, atopic eczema, periodonditis and inflammatory bowel disease (IBD). IBD is a chronic inflammatory disease of the gastrointestinal tract. It is divided into two main sub diseases; Crohn's disease (CD) and ulcerative colitis (UC).Their classification is based on clinical, endoscopic, histological and pathological parameters, including the distribution of inflammation in the colon and/or small intestine. There is a significant increased risk of developing either forms of IBD for relatives to diseased individuals[1].
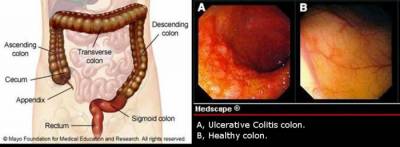
The project aims to identify and characterize epigenetic modifications such as altered DNA methylation profiles that are indicative of or contributory to the pathophysiology of inflammatory bowel disease (IBD). Such methylation profiles will represent the first link between the predetermined genetic basis of susceptibility and flexible environmental factors. By integrating different technologies such as genome wide mRNA expression profiling together with high throughput methylation profiling of the epigenome, a correlated IBD specific DNA methylation/ expression profile will be generated. Discordant monozygotic twins represent an ideal cohort for monitoring association between epigenetic and transcriptional differences and the transcription/ methylation levels will be analysed in mucosa biopsies.
DNA methylation and disease
DNA methylation is the most common epigenetic modification and occurs nearly exclusively at cytosines in CpG dinucleotide enriched areas, which are often located in the promoter region of many genes. For instance, hypermethylation of promoter regions often results in gene silencing, while promoters of transcriptionally active genes are typically hypomethylated[2]. Thus a direct link between methylation and the functional phenotype of a cell, organ or even organism can be postulated.
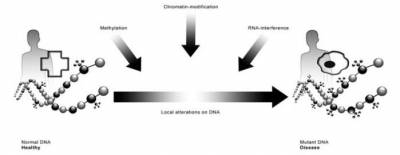
Disruption of methylation patterns is a characteristic feature in cancer where hypermethylation of tumor suppressor genes can lead to tumor progression and global hypomethylation may lead to activation of oncogenes [3, 4]. Methylation positions have been identified that correlate with cancerdiabetes type II[5], arteriosclerosis[6], rheumatoid arthritis[7] and neurodegenerative diseases[8]. Methylation at other positions has been shown to correlate with age, gender, nutrition, drug use, and probably a whole range of other environmental triggers[9, 10]. DNA methylation is one of the basic mechanisms that control transcription. It plays a role in chromosome X-inactivation, in silencing of tumour genes, in imprinting, and in many other fields and diseases[11] as described above. Patterns of DNA methylation reflect the pattern of gene activity within a given cell type and functional cellular state. A recent publication indicates that homozygotic twin's global methylation profiles diverge over time [12], which indicates regulated differential methylation possibly due to lifestyle differences. Understanding DNA methylation changes and their correlation to gene expression patterns may therefore provide important novel insights into the complex pathophysiology of inflammation. As DNA methylation may be the most important flexible genomic parameter that can change genome function under an exogenous influence, it most likely provides the main link between the genetics of disease, and the environmental components that are widely acknowledged to play the decisive role in the aetiology of all inflammatory diseases in the focus of biomedical research today. New techniques have recently been developed which enable genome-wide methylation profiling.
One promising technique is methylated DNA immunoprecipitation MeDIP [13, 14] this method relies on immuno recognition to detect and precipitate methylated DNA, that later will be hybridized to a DNA array. Alternative methods to fractionate hyper- or hypo-methylated DNA sequences, include the use of methylation-sensitive restriction enzymes[15]. The disadvantage however with this method is that it can present a sequence bias. Thus far two studies have been performed in cancer, using the MeDIP/DNA array techniques [13, 16]. DNA methylation differences however have never been investigated on a global basis in inflammation and this information, which is now accessible will yield information about the role of methylation in transcription generally[17]. IBD is a suitable model disease since it has a clearly demonstrated polygenetic component, is also influenced by environmental factors, has well defined phenotypes, and shares many similarities with other complex diseases. The biomedical importance of a disease-focused approach is emphasised by the fact that only well characterized phenotypes can be correlated to methylation patterns. Discordant monozygotic twins represent an ideal cohort for monitoring association between epigenetic and transcriptional differences and the methylation levels since they have identical genomes but differ in disease status. Levels of both gene expression and methylation will be analysed in mucosa biopsies. Using intestinal mucosa biopsies is of great advantage as opposed to e.g. blood, since it is the tissue in which disease occurs, and it is accessible to environmental influences: The human intestine represents the largest interface between an individual and the environment.
Methods
The IBD methylation profile will be generated from an integrated genome wide mRNA expression/ DNA methylation profiling analysis in mucosal tissues. mRNA expression profiling will be performed by Professor S. Schreiber's research group at U. of Kiel. Both analyses will be done on IBD discordant monozygotic twin pairs and healthy monozygotic twin pairs.
The global methylation profiling will be performed using high resolution tiling-path arrays. The analysis will be performed on mucosa biopsies from the monozygotic twins. In brief, sonicated genomic DNA is incubated with antibodies against methylated DNA. Methylated genomic fragments are separated by immuno-precipitation. This fraction is labeled with Cy3 and mixed with equal amounts of un-enriched DNA labeled with Cy5 and subsequently hybridized to the tiling path array. This method will determine methylation levels across the epigenome.
 Close
Close


