Scientists reveal variability as key mechanism behind adaptability of immune cells
2 March 2017
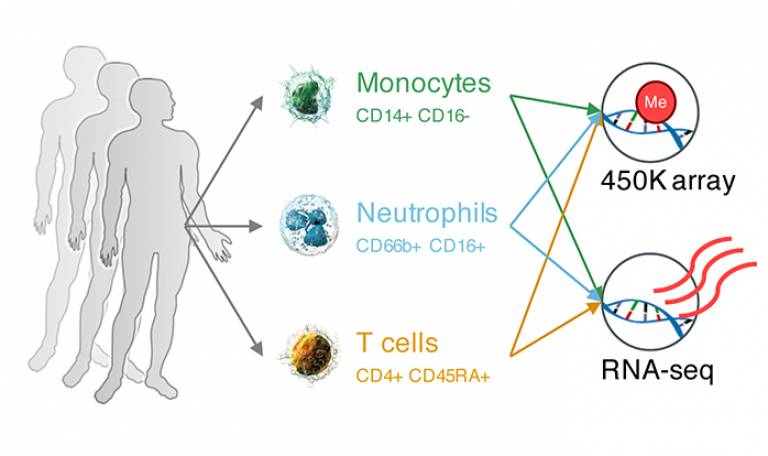
Scientists from UCL, the Wellcome Trust Sanger Institute, and the University of Cambridge have identified differences in transcriptional and epigenetic variation between immune cell types that could provide vital clues to understanding how these cells adapt and respond to changing environmental conditions. These findings will inform research into how this variation can be influenced to benefit patients with immune-related diseases such as type 1 diabetes and cancer.
Using purified blood cells from 125 healthy people, the research team focused on comparing three types of immune cells: monocytes, neutrophils and T cells – chosen due to the crucial role they play in the body’s immune response and their relative abundance in blood. The team looked at how much the expression and DNA methylation in each type of cell varied between individuals, rather than only focusing on the overall average expression and methylation differences between cell types.
“We know that variation is relevant to disease,” explains Dr Simone Ecker, Research Associate at UCL Cancer Institute and lead author of the study. “For example, we know that in cancer – a very heterogeneous disease – there is generally high variability. In this study, we wanted to investigate healthy cell populations to understand to what extent variability is present there and what function it might serve. Elucidating these processes in healthy cells will help us better understand disease states.”
When the study team directly compared the three cell types to each other, they found that gene expression levels in neutrophils varied greatly between individuals compared to monocytes and T cells, where gene expression was more consistent across the study participants. Similar patterns were observed for DNA methylation.
“We thought that this increased variation in neutrophils reflects the fact that these cells are the immune system’s ‘first responders’. They are highly mobile and migrate to sites of infection and react quickly to pathogens,” says Dr Dirk Paul from the University of Cambridge, who co-led the study. “Neutrophils need to be highly ‘flexible’ in how they respond, so the variability has a function, it enables this adaptability,” adds Dr Ecker.
But while this increased variation in neutrophils serves a valuable purpose in maintaining immunity, enhanced variability in diseases such as cancer can be a threat, because cancer cells exploit adaptability to grow and evade treatment.
“Previous studies have shown that high variability correlates with the development and progression of disease,” explains Dr Ecker. “Research has also shown that high variability at different levels increases the aggressiveness of cancer. Our previous work on leukaemia supports this concept. We looked at two clinical subtypes of leukaemia, one of which is indolent – patients do often not receive treatment and continue their lives almost normally; the other type is aggressive and survival rates are much worse. If you investigate mean gene expression differences between the two types of disease, you will not see anything striking, but when you compare expression variability between these two groups, the aggressive type shows significantly increased variability.”
Deepening our understanding of this variability, in particular at the single-cell level, will be critical in helping to develop improved ways of targeting cancer and to tailor treatment to patients. Further research in this area has the potential to reveal targets that might reduce variability in cancers and immunological diseases without compromising the function of the immune system.
This study was undertaken in collaboration with the BLUEPRINT epigenome project and was supported by the UCLH NIHR Biomedical Research Centre.
Further information
- Research paper: ‘Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types’ Genome Biology
- Dr Simone Ecker academic profile
- Medical Genomics Group - Professor Stephan Beck
- Integrative Human Genomics Team - Dr Dirk Paul
- BLUEPRINT Consortium
- Opinion article: Epigenetic variation taking centre stage in immunological research Dr Simone Ecker and Professor Stephan Beck
Related news
New research leads the way in epigenomic studies
Research led by UCL and Queen Mary University of London (QMUL) scientists has identified epigenetic changes in three types of immune cell that could contribute to the development of type 1 diabetes. These changes were only found in patients with diabetes, opening the door to future biomarker development.

BLUEPRINT celebrates major manuscript release
European epigenomes research project, BLUEPRINT, today announced the release of a collection of 26 publications in Cell, Cell Press-associated and other high-impact journals. The papers are part of a package of 41 publications by the International Human Epigenome Consortium (IHEC) of which BLUEPRINT is a member.
 Close
Close

