The research in our laboratory focuses on the processes that establish cell diversity and pattern in the early embryo. We ask the questions: how do cells in the embryo know what fates to adopt, at the right positions and at the right time? What mechanisms ensure that the correct proportions of cells are allocated to different organs?
Currently, the projects in the lab fall into four major headings:
2. What mechanisms are responsible for inducing the early nervous system?
3. How is the early nervous system subdivided into forebrain, midbrain, hindbrain and spinal cord?
We are particularly interested in discovering mechanisms that represent general principles in development, and therefore follow a multi-disciplinary approach. We choose (or if necessary, develop) techniques that will help us best to answer the questions being asked, rather than being wedded to any particular set of techniques. We do not define our questions based on specific genes, but rather based on the biological event we are trying to understand - we first define the biological process and then try to establish which genes are important for that process. Finally, although much of our research uses chick embryos (because they are easy and cheap to obtain at precise stages of development, because it is easy to manipulate cells, and because a lot is already known about how they develop), we are also not wedded to this as an experimental system. Current projects use chick, quail, frog and mouse embryos, and we are also collaborating with other labs using yeast, flies and fish.
1. How do higher vertebrate embryos establish their polarity, and what mechanisms coordinate cell movements with gene expression?
Early during development, the embryo learns which of its ends will start to form the future axis. Most of what we know about this process comes from studies in the fly and the frog. In both of these organisms, polarity is established initially by the localisation of maternal determinants (which may be RNAs or proteins) to one or the other end of the cytoplasm of the fertilised egg. As cleavage begins, cells forming in different positions inherit different sets of these determinants, and their fates will be fixed accordingly. This mechanism is facilitated by the fact that both flies and frogs simply subdivide the initial egg volume among the forming cells, so that there is essentially no growth (and hence, no dilution of the determinants) during the early phases of development. A second feature of these embryos is that the transcription of zygotic genes does not begin until several cleavages have occurred and the embryo contains many cells (in the frog, after about 10-11 cleavages, at the "mid-blastula transition"). A consequence of this mechanism is what is known as "mosaic development": for example, if the cells of an early frog embryo say, at the 8 cell stage, are separated from each other, different cells will give rise to different structures, and few (if any) will generate a whole embryo. Higher vertebrate embryos, such as birds and mammals, are rather different. First, they increase greatly in volume from the very earliest stages of development. Second, there is no "mid-blastula transition", and zygotic gene expression begins very soon after fertilisation. Third, these embryos are "regulative" (rather than mosaic): a chick embryo at the late blastula stage (containing as many as 20,000 cells) can be cut into many fragments, all of which will generate a complete, but smaller embryo, as demonstrated by Nelson Spratt in 1960. In the frog, two determinants have been demonstrated to be particularly important in setting up initial polarity: the activin/Vg1 pathway in the vegetal half of the embryo, and activation of the Wnt/β-catenin pathway at the dorsal side. The quadrant where they overlap becomes a special inducing region, called the Nieuwkoop centre. This region is defined as a group of cells which have the ability to induce the development of a second inducing centre, called Spemann's organizer (which in turn induces the nervous system - see project 2 - and also sets up dorsal/ventral polarity in the mesoderm), but without contributing cells to the organizer.
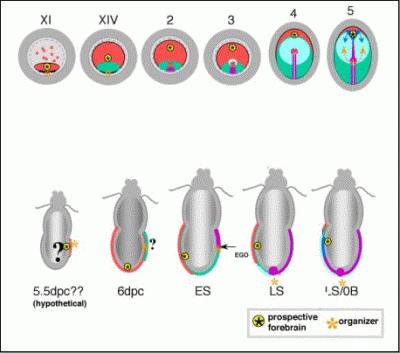
Stages of chick (upper) and mouse development
Is the axis of higher vertebrate embryos established by similar mechanisms? Despite their rather different mode of development, it appears that the embryos of higher vertebrates use similar signalling pathways to specify the orientation of the axis. We have shown that a region analogous to the Nieuwkoop centre exists in chick embryos (Bachvarova et al., 1998), and that it is located in the posterior margin of the embryonic disk. Vg1 is specifically expressed in this region, and misexpression of Vg1 at any other point of the margin can initiate the formation of a second embryonic axis (Shah et al., 1997). However, this misexpression must be done in the margin - other regions of the embryo cannot respond. We have now shown that the reason for this is that Wnt8C is expressed all around the margin (Skromne and Stern, 2001). Therefore, as in amphibians, the embryonic axis is initiated in a region where Vg1 and Wnt activities overlap.
Vg1+Wnt8C induce other signalling molecules as part of a cascade. One of these is Nodal (a member of the TGF-beta family) which, probably in cooperation with FGFs, is responsible for inducing the primitive streak as a site of mesoderm and endoderm formation. However, Nodal (which is expressed in the epiblast) cannot act because it is inhibited by its antagonist, Cerberus, expressed in the underlying hypoblast (Bertocchini and Stern, 2002). Primitive streak formation is initiated only after the hypoblast becomes displaced by another tissue, the endoblast, which does not expressed Cerberus. This mechanism therefore controls both the timing and the position of primitive streak formation.
Now many important questions remain to be answered. For example, since the chick embryo is regulative (a new axis forms spontaneously if the site of normal axis formation is removed), what positions Vg1 and Wnt at a new site? Since any point around the circumference of the normal embryo is able to form an axis, what ensures that only one axis forms during normal development? Several observations point to the existence of inhibitors of axis formation, which we are now trying to identify.
Coordination of cell movements and gene expression. During the process of axis formation, and subsequently during gastrulation, the cells of the embryo are not static in defined positions, but move around rather a lot. At the same time, the expression of many genes (including Vg1 and Wnt8C) is restricted to specific locations in the embryo. How can these two observations be reconciled? We have shown that as cells move around, they turn on and off the expression of genes appropriate to their current locations - therefore at least in early embryos, gene expression patterns are not indications of cell lineage history but rather of the current position of the cells (Joubin and Stern, 1999). There must therefore be positional signals that instruct cells of their current positions in the embryo, and we have shown that an interplay between inducers and inhibitors is involved even after the axis has started to form. The inducers include, again, Vg1 and Wnt, and the inhibitors include Anti-dorsalising Morphogenetic Protein (ADMP, another member of the TGFβ family like Vg1) and BMP-2, -4 and -7 (which are also related to TGFβ) (Joubin and Stern, 1999). We are now trying to visualise these processes in real time in living embryos, using the promoter of an endogenous gene (goosecoid) expressed specifically in the organiser to drive expression of a fluorescent reporter. This allows us to film cell movements at the same time as to see the onset of expression of the goosecoid gene, and to combine this with manipulation of cells or of the expression of other genes to study how these events are regulated in vivo.
2. What mechanisms are responsible for inducing the early nervous system?
The previous section addressed questions about how the polarity of the early axis is established, and the mechanisms that position the organiser in its correct location. One of the reasons why understanding these mechanisms is important is that the organiser plays a critical role in the subsequent stages of development. Among its functions is the phenomenon of neural induction: the organiser emits signals that instructs cells nearby to become nervous system, while cells further away that do not receive these signals develop into skin. What are these signals?
In amphibians, a large body of published evidence is generally interpreted as implicating secreted factors of the BMP family in this process: BMPs appear to act as neural inhibitors and epidermal (skin) inducers. The organiser emits BMP antagonists (such as Chordin, Noggin, Follistatin and other proteins), which inhibit BMP activity in the vicinity of the organiser, releasing these cells from the neural inhibition exerted by BMP. This view is generally known as the "default model". However, evidence from chick embryos and close scrutiny of some of the amphibian literature (reviewed by Streit and Stern, 1999) suggests that the events of neural induction may be rather more complex, involving several steps.
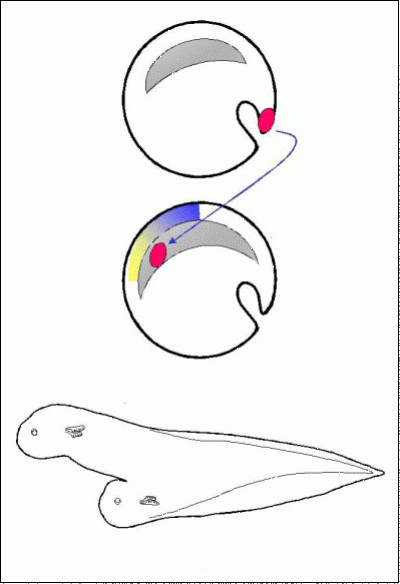
Spemann's & Mangold's experiment (1924) For example, misexpression of BMP antagonists in chick embryos, under identical conditions as those that allow a grafted organiser to induce a complete axis, do not result in cells acquiring a neural fate (Streit et al., 1998). However, when an organiser is grafted, removed after 5 hours (which is not sufficient t ime for it to induce a nervous system) and replaced by Chordin-secreting cells, cells now maintain the expression of an early neural marker, Sox3 (Streit et al., 1998). This finding suggests that signals from the organiser other than BMP antagonists are required for cells to become responsive to the BMP antagonists. However, even more steps must be involved, because even in this experiment later neural markers (like Sox2, or regionally-specific markers, or markers for terminally differentiated neurons or glia) are never expressed. We started by analysing the state of cells that have been exposed to signals from the organiser for 5 hours. Using new technology that allows construction and screening of cDNA libraries made from single cells, we conducted a differential screen between chick cells that had been exposed signals from a grafted organiser for 5 hours and cells that had not. A total of 15 differentially expressed genes were identified, of which 11 are novel sequences. The first of these genes to be studied in detail was named ERNI, which encodes a novel coiled-coil protein (Streit et al., 2000). Reassuringly, this gene is expressed in the normal embryo in cells destined to form the neural plate, but only transiently. By the time the neural plate starts to form, ERNI expression is turned off. Having this early marker for cells that have received the initial signals of neural induction allowed us to search for the signals themselves. Of many factors tested, the only one that can induce ERNI expression with a similar time-course as a grafted organiser is a member of the Fibroblast Growth Factor family, FGF8. Moreover, FGF8 is expressed in cells that will later give rise to the organiser and later in the organiser itself (Hensen's node), and inhibition of FGF signalling prevents grafted organisers from inducing both ERNI expression and a neural plate (Streit et al., 2000). This is good evidence that FGF is an early signal required for neural induction, and the expression of FGF8 and ERNI suggest that the early steps of this process occur very early in development, even before the beginning of gastrulation.
But FGFs have many functions in development. Even at this early stage, they are involved in mesoderm induction as well as in the initiation of neural induction, events that happen almost simultaneously at the beginning of gastrulation yet their outcomes are incompatible with one another (a cell cannot become both mesoderm and neural). How do cells decide how to respond to these signals? An answer came from one of the other genes isolated in the screen, which we have named Churchill. This encodes a novel zinc finger transcriptional activator, one of whose targets is Smad-interacting-protein-1 (Sip1). In turn, Sip1 inhibits transcription of a factor required for mesoderm formation, brachyury, which is also an "immediate-early" target of FGF. Like ERNI, Churchill is induced by FGF8 but not as quickly - 4-5 hours are required. Consistent with this, Churchill begins to be expressed only towards the end of gastrulation in the normal embryo. Therefore Churchill seems to act as a delaying mechanism, separating two functions of FGF: an early requirement for mesoderm induction and a slightly later role in initiating neural plate development (Sheng et al., 2003). We are continuing the study of the remaining genes isolated from our screen, which are also revealing more interesting features of the earliest steps in the induction of the nervous system. But the experiments that led to this screen (organiser grafts, removal and replacement by BMP antagonists), as well as co-expression of FGF and BMP antagonists, suggested that signals other than these two are also required (Linker et al., 2004). To start to identify them, we have recently conducted a screen using technology developed by the Genetics Institute in Boston, which allows the construction of cDNA libraries enriched for secreted products. We made a library from cDNA isolated from the organiser at a stage when it can induce a complete nervous system, selected it for putative secreted products, and analysed the embryonic expression of all of the likely inserts. We identified 3 distinct novel cDNAs encoding putative secreted factors that are expressed appropriately, in the correct place (Hensen's node and/or primitive streak) and correct time (early to mid-gastrulation stages) to be involved in neural induction. We are currently investigating whether any of these encode the missing factor(s). This project illustrates our favourite approach to studying developmental problems. Starting with a defined biological problem, we search for the genes that might be involved to then study their functions in the chosen problem (top-down approach), rather than starting from the gene to establish its function (bottom-up approach).
3. How is the early nervous system subdivided into forebrain, midbrain, hindbrain and spinal cord?
The preceding project addressed the question: how is the nervous system induced? But grafting an organiser to a new site generates not just non-specific neural cells, but a fully patterned, organised nervous system. It is this classical finding that led to the name "organiser" for this region of the embryo. How does the organiser generate the initial subdivisions of the nervous system?
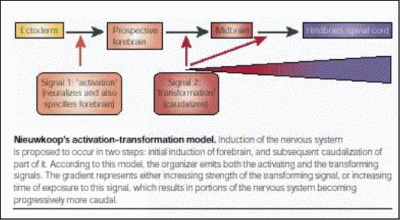
This problem has preoccupied embryologists for almost a century, and many models have been proposed. One of these, which developed from findings made initially by Hans Spemann and his colleagues in the early 1930's, postulated the existence of separate organisers for the head, trunk and tail. Another, proposed by Pieter Nieuwkoop in the 1950's, suggested that all of the neural tissue initially induced ("activation") by the organiser has forebrain character, but that later signals also emanating from the organiser or from its descendant cells "transform" some of this neural tissue to more posterior (caudal) fates. Different types of experiments performed in the last 5 decades are consistent with one or the other model, but none clearly distinguished between them. Recently, evidence obtained in the mouse embryo has been interpreted to support the former model - it was suggested that a "head organiser" resides outside of the classical organiser region (Hensen's node), in a tissue that will only contribute to extraembryonic membranes, the Anterior Visceral Endoderm (AVE).
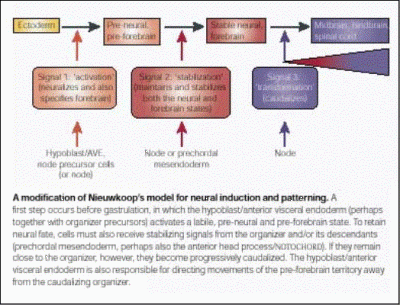
However, this interpretation does not easily explain some findings, for example, why does a graft of Hensen's node in the chick embryo generate a complete axis that includes a forebrain, even when placed in a region that has never been exposed to the chick equivalent of the AVE (the hypoblast)? Recently, we have obtained evidence that reconciles the two opposing models, while at the same time strongly supporting the latter one. Grafts of the hypoblast, while being unable to induce either a nervous system or a head directly, transiently induce the expression both of early neural (such as ERNI and Sox3) markers and of a gene which later becomes restricted to the forebrain, the transcription factor Otx2. However, unless an organiser (Hensen's node) is also grafted, expression of all of these markers is lost. Some evidence suggests that the signal responsible for this unstable induction is an FGF, (perhaps FGF8, which is expressed in the hypoblast at this stage): misexpression of FGF8 induces transient expression of all three markers with the same time course as a grafted hypoblast (Streit et al., 2000; Foley et al., 2000).The hypoblast also plays an additional role. In the early 1930's, Waddington demonstrated that the hypoblast can influence the orientation of the embryonic axis. When the hypoblast is rotated with respect to the rest of the embryo, the axis that develops bends as if trying to align itself with the rotated hypoblast. We have shown (Foley et al., 2000) that the reason for this is that the hypoblast regulates the direction of cell movements in the other tissues (epiblast and mesoderm). When cells that would normally contribute to the forebrain are tracked in this experiment, they still contribute to the forebrain, even though this structure now develops in a different position. This experiment shows that, rather than inducing the forebrain, the hypoblast regulates cell movements that position this region in its appropriate place (Foley et al., 2000).Taken together, these experiments argue against the idea that separate organisers are responsible for inducing the head, trunk and tail portions of the nervous system. We therefore proposed a model (Foley et al., 2000; Stern, 2001) that reconciles the differences between the >classical views of how the neural axis is initially patterned. An initial event similar to the "activation" step of Nieuwkoop's model induces transient and unstable expression of early neural (Sox3, ERNI) markers, as well as of the future forebrain marker Otx2. But development of the forebrain requires at least one additional signal, which stabilises the first. A third signal (or set of signals), equivalent to the second ("transformation") step of Nieuwkoop's model, also stabilises the neural state as well as transforming it to more posterior character.We are now testing this model directly and trying to identify the signals responsible.
 Close
Close

