The UCL Nanostring Facility has been established to provide access to the nCounter technology for the research community. The facility owes its origins to a UCL Platform Technologies Initiative Grant and it is managed by the GEE Department.
COVID-19: The UCL Nanostring facility is open for external service run by our staff members, no training is available.
The nCounter System
The nCounter system from Nanostring Technologies is a direct, reliable and highly sensitive multiplexed measurement of nucleic acids (DNA and RNA) based on a digital barcode technology.
The nCounter system is fully automated and uses fluorescently labelled reporter probes, called 'codesets', and single molecule imaging to detect and count hundreds of molecules in a single reaction. The system is very resistant to poor quality of the RNA samples and it is perfectly suited for RNA extracted from FFPE (formalin-fixed, paraffin-embedded) samples.
The nCounter technology has sensitivity and specificity comparable to real time PCR (RT-PCR) and it is faster than NGS. This technology takes advantage of the well-understood nucleic acid hybridization reaction and no enzymatic reactions are involved, no reverse transcription or DNA amplification is necessary.
For any enquiry about the nCounter service at UCL please use the following email address: nanostring@ucl.ac.uk.
| Paola Oliveri | Head of Facility |
| Fraser Simpson | Technical Staff |
| Wendy Hart | Technical Staff |
For Codeset design and ordering, please contact Nanostring directly: Xavier Tait (email) or xtait@nanostring.com
The Nanostring technology is perfect tool for validation of NGS and microarray data; for quantification of multiple genes from limited amount of material; for pathway analysis, expression analyses of defined gene sets, for biomarker validation and more.
Gene expression analysis
Quantification of specific transcripts in a pool of total RNA using either the preassembled and ready-to-use panels or the flexible custom codesets. Compatible with Total RNA, FFPE, cell lysate, plasma, serum and more.
miRNA profiling
Quantification of micro-RNA using ready panels from Human, Mouse and Rat that contain up to 800 targets or à la carte option with up to 50 custom selected miRNA probes. Compatible with miRNA from serum, plasma, FFPE, tissue, cells and exosomes
miRGE™ (miRNA & mRNA) analysis
mRNA and miRNA simultaneous quantification. Compatible with total RNA from serum, plasma, FFPE, tissue, cells, and exosomes.
Vantage 3D
Simultaneous quantification of combinations of DNA, RNA, and protein targets up to 800-plex from a single sample.
The UCL Nanostring facility is open to every University, Research Institute and Private Company and provides the following types of service:
- Full nConter service: all the steps from sample quality control to initial data analysis are carried out by UCL experienced staff.
- nCounter machine training: a researcher is trained in using the nCounter machine for the first 12 samples. After he/she will be given access to the machine to run other samples. This service is available for large projects. (This Service is temporally suspended due to COVID Restrictions)
- Data analysis: background correction and normalization are provided in a excel format.
- Data analysis training: a person is trained in data analysis using excel.
For off-the-shelf preassembled Probe Sets that include nCounter Master Kits with all of the consumables and reagents to perform the assay, please contact Nanostring directly.
For custom projects, probes for a Codeset are designed by nanostring bioinformaticians, on the basis of RefSeq ID's or Fasta sequences. This format is available for up to 800 target sequence and for 48 or 96 reactions, or multiples.
An nCounter Codeset contains positive controls (10 RNA spiked in the Codeset) and negative controls (7 probes against alien sequences) that are used for internal QC, calibration and background evaluation.
Similar to RT PCR, probes for normalization genes must be included in the Codeset. These must be selected based on prior knowledge.
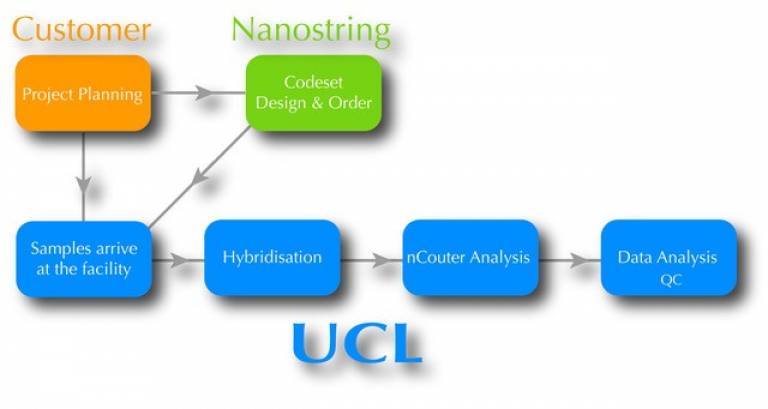
How to start the process
If this is the first time using the service please contact the facility in advance by email regarding your project ideas. We can answer any questions that you may have, and advise you on what technique would best suit your needs. Tell us about your sample type, species and objectives.
Once agreed on the type of service, please fill out the Registration Form. You will take care to order the necessary codeset directly from nanostring.
How to prepare the samples
A minimum amount of RNA/DNA sample is required for the nCounter quantification. For more details please see table below and download the
.
Sample preparation summary
| Application | Starting Material | Recommended Sample Concentration | Assay Input Amount | Total Amount Required per Assay* |
| Gene Expression | Total RNA | 20 ng/µL - 100 ng/µL | 100 ng | 150 ng |
| Cell lysate | 2,000 - 10,000 cells/µL | 10,000 cells | 15,000 cells | |
| FFPE RNA | 20 ng/µL - 100 ng/µL | 100 ng | 150 ng | |
| miRNA | Purified total RNA only | 33 ng/µL - 100 ng/µL | 100 ng | 150 ng |
| FFPE RNA | 33 ng/µL - 100 ng/µL | 100 ng | 150 ng | |
| CNV | Purified Genomic DNA only | >85 ng/µL | 600 ng | 650 ng |
*When shipping samples to NanoString, please provide an additional 50 ng of material to account for dead volume pipetting and additional sample QC as necessary. Material for technical replicates can be provided in a single tube or plate well.
Quality control of RNA samples can be carried out at extra cost using the BioAnalyzer. In this case we have to request an amount of at least 300 ng RNA (between 20 - 100 ng/uL). If samples fail to meet incoming quality checks they will be rejected with a given reason.
In case you want to readout a cartridge, please fill in the Cartridge Submission Form.
NOTE: All prices are subject to change without notice.
The customer will have to pay a service fee for all consumables, handling of samples, data generation and processing. Prices vary depending on the type of experiment and service provided by the facility.
Please cite our service in the acknowledgements section of your publication in case we have just performed the standard service. The following statement may be used:
"We thank the UCL Nanostring Facility for providing the nCounter system and related services."
Is the price per sample or per experiment?
- Our pricelist provide price/sample or price/cartridge (= 12 samples).
Do you extract the RNA from my sample?
- No our facility do not provide RNA extraction service.
Do you provide analysis of the data?
- We provide an initial data analysis, QC and cartridge normalization, but we do not provide any advanced data analysis.
How long does it take to have the result?
- It depends on how busy is already the service. The workflow for 12 samples takes 48 hours.
Can I be trained to use the Nanostring machine?
- Yes we train experience researches (e.g. postdoc) only for projects larger of 48 samples. (This Service is temporally suspended due to COVID-19 restrictions)
Will we be expected to pay a separate fee to Nanostring for ordering an RNA codeset - or is this included in the quoted price?
- UCL quote includes only the experiment running fee. The reagents need to be purchased directly from Nanostring.
Is there a minimum number of samples to analyse per run?
- No, however we suggest to use all the 12 well in a cartridge, which otherwise will be lost.
Is your price per sample for up to 800 genes? Or does the price vary depending on the number of genes analysed?
- The price/sample indicate in the pricelist, does not change with the number of genes present in the codeset.
How much RNA would I need to provide to run 30-50 genes?
- The RNA amount required is the same for 30 or for 800 genes and depends on the type of essay.
In what format will I receive the data? Has any processing been done?
- We send the data in 2 formats: excel with QC and background control performed, and RCC file that can be load directly into the nSolver free software provided by Nanostring.
Is there a waiting list for running samples?
- It depends on the moment of the year, generally yes.
What if a gene doesn't have a RefSeq ID for making codesets?
- You can provide to Nanostring the sequence in Fasta format.
 Close
Close


