Global Lactase Persistence Association Database GLAD
Lactase persistence (LP) is the ability of adult humans to digest the milk sugar lactose. LP is a dominant Mendelian trait that has been a subject of extensive genetic, medical, and evolutionary research.
This is an online database resource presenting LP phenotypic and genotypic allele frequency data: interpolated LP frequency maps are shown, focussing on the Old World.
We encourage researchers to send us new LP phenotype / genotype data to Professor Dallas Swallow in an Excel format using headers identical to those in the tables linked below.
Please make sure that the submitted phenotype data conform to the following criteria:
- Random sampling
- Known ethnicity and sampling location
- Indigenous population (preferably, but please alert us and supply details, if not)
- No hospital patients or individuals self reporting with gut problems
Latest Maps
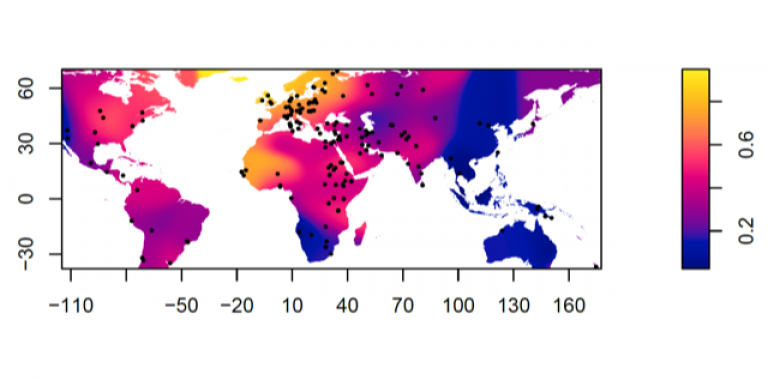
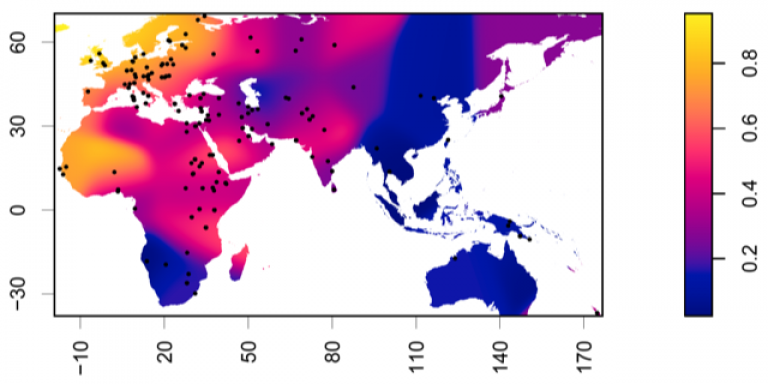
Distribution of Lactase Persistence phenotype
Dots represent test locations. Colours and colour key show LP frequency distribution represented by surface interpolation. Ingram,C.I, Montalva, N and Swallow, D.M. 2022 'Lactose Malabsorption’, in Advanced Dairy Chemistry, Volume 3: Lactose, Water, Salts and Minor Constituents McSweeney et al eds. ISBN 978-3-030-92584-0. See data links below.
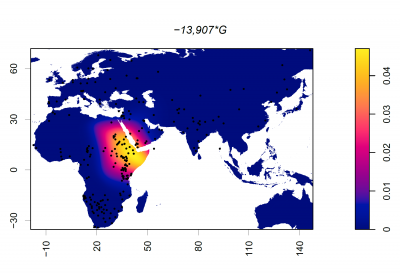
Frequency of 13907*G (rs41525747)
Dots represent collection locations. Colours and colour key show the allele frequency distribution represented by surface interpolation. Taken from Ingram,C.I, Montalva, N and Swallow, D.M. 2022 'Lactose Malabsorption’, in Advanced Dairy Chemistry, Volume 3: Lactose, Water, Salts and Minor Constituents McSweeney et al eds. ISBN 978-3-030-92584-0. See data links below.
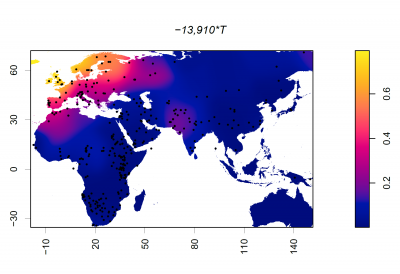
Frequency of 13910*T (rs4988235)
Dots represent collection locations. Colours and colour key show the allele frequency distribution represented by surface interpolation. Taken from Ingram,C.I, Montalva, N and Swallow, D.M. 2022 'Lactose Malabsorption’, in Advanced Dairy Chemistry, Volume 3: Lactose, Water, Salts and Minor Constituents McSweeney et al eds. ISBN 978-3-030-92584-0. See data links below.
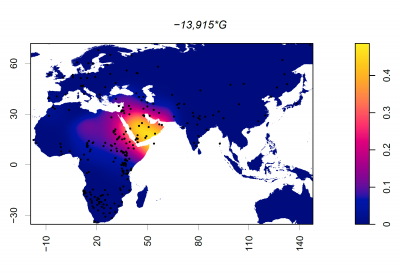
Frequency of 13915*G (rs41380347)
Dots represent collection locations. Colours and colour key show the allele frequency distribution represented by surface interpolation. Taken from Ingram,C.I, Montalva, N and Swallow, D.M. 2022 'Lactose Malabsorption’, in Advanced Dairy Chemistry, Volume 3: Lactose, Water, Salts and Minor Constituents McSweeney et al eds. ISBN 978-3-030-92584-0. See data links below.
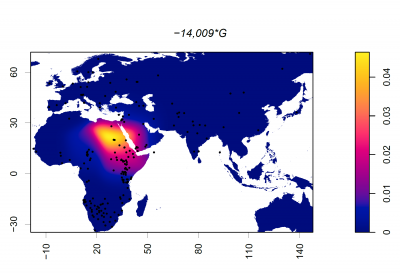
Frequency of 14009*G (rs869051967)
Dots represent collection locations. Colours and colour key show the allele frequency distribution represented by surface interpolation. Taken from Ingram,C.I, Montalva, N and Swallow, D.M. 2022 'Lactose Malabsorption’, in Advanced Dairy Chemistry, Volume 3: Lactose, Water, Salts and Minor Constituents McSweeney et al eds. ISBN 978-3-030-92584-0. See data links below.
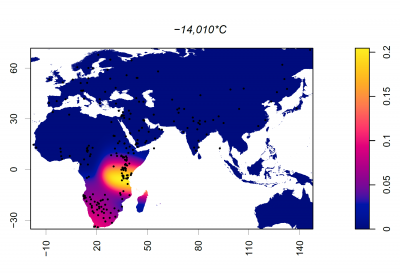
Frequency of 14010*C (rs145946881)
Dots represent collection locations. Colours and colour key show the allele frequency distribution represented by surface interpolation. Taken from Ingram,C.I, Montalva, N and Swallow, D.M. 2022 'Lactose Malabsorption’, in Advanced Dairy Chemistry, Volume 3: Lactose, Water, Salts and Minor Constituents McSweeney et al eds. ISBN 978-3-030-92584-0. See data links below.
Data
| Database | Version | Reference |
LP Genotypes
Old World | 2012 - DOWNLOAD | Itan et al 2010 |
| LP Phenotypes | 2012 - DOWNLOAD | Itan et al 2010 |
LP Genotypes
Old World | 2017 - DOWNLOAD | Liebert et al 2017 |
LP Genotypes
Old World | 2020 - DOWNLOAD | Ingram,C.I, Montalva, N and Swallow, D.M. 2022 'Lactose Malabsorption’, in Advanced Dairy Chemistry, Volume 3: Lactose, Water, Salts and Minor Constituents McSweeney et al eds. ISBN 978-3-030-92584-0 |
LP Phenotypes
with Americas | 2020 - DOWNLOAD | Ingram,C.I, Montalva, N and Swallow, D.M. 2022 'Lactose Malabsorption’, in Advanced Dairy Chemistry, Volume 3: Lactose, Water, Salts and Minor Constituents McSweeney et al eds. ISBN 978-3-030-92584-0 |
LP Ancient
Genotypes | 2022 -
DOWNLOAD | Evershed, R.P., Davey Smith, G., Roffet-Salque, M. et al. Dairying, diseases and the evolution of lactase persistence in Europe. Nature (2022). Data taken from v44.3 of the Allen Ancient DNA Resource (accessible at: https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data). Single alleles (for read depth one) and maximum likelihood genotypes (for read depth above one) were obtained from pileups on BAM files for the set of individuals shown in Figure 2 of Evershed et al. As described in Evershed et al., rs4988235 genotypes for individuals PEN001_real2, Klei10.SG, and Iboussieres25-1 were manually set to the GG, as presence of the derived LP allele is believed to be due to post mortem damage. |
Credits
Maps
Nicolas Montalva.
Database
Anke Liebert, Yuval Itan, Bryony Leigh Jones, Catherine Ingram, Dallas Swallow, Mark Thomas, Nic Montalva, Yoan Diekmann.
Corrections
Thanks to Favia Domizia Nardi, Laure Segurel, Pavel Grasgruber, and several others for supplying corrections.
Last update
February 2023 by Dallas Swallow and Mark Thomas
 Close
Close