Publications
CANDELA paper on facial features published in Nature Communications:
Summary from the article:
We report a genome-wide association scan for facial features in ~6,000 Latin Americans. We evaluated 14 traits on an ordinal scale and found significant association (P values<5 × 10−8) at single-nucleotide polymorphisms (SNPs) in four genomic regions for three nose-related traits: columella inclination (4q31), nose bridge breadth (6p21) and nose wing breadth (7p13 and 20p11). In a subsample of ~3,000 individuals we obtained quantitative traits related to 9 of the ordinal phenotypes and, also, a measure of nasion position. Quantitative analyses confirmed the ordinal-based associations, identified SNPs in 2q12 associated to chin protrusion, and replicated the reported association of nasion position with SNPs in PAX3. Strongest association in 2q12, 4q31, 6p21 and 7p13 was observed for SNPs in the EDAR, DCHS2, RUNX2 and GLI3 genes, respectively. Associated SNPs in 20p11 extend to PAX1. Consistent with the effect of EDAR on chin protrusion, we documented alterations of mandible length in mice with modified Edar funtion.
Link: http://www.nature.com/ncomms/2016/160519/ncomms11616/full/ncomms11616.html
CANDELA paper on hair variation published in Nature Communications:
Summary from the article:
We report a genome-wide association scan in over 6,000 Latin Americans for features of scalp hair (shape, colour, greying, balding) and facial hair (beard thickness, monobrow, eyebrow thickness). We found 18 signals of association reaching genome-wide significance (P values 5 × 10−8 to 3 × 10−119), including 10 novel associations. These include novel loci for scalp hair shape and balding, and the first reported loci for hair greying, monobrow, eyebrow and beard thickness. A newly identified locus influencing hair shape includes a Q30R substitution in the Protease Serine S1 family member 53 (PRSS53). We demonstrate that this enzyme is highly expressed in the hair follicle, especially the inner root sheath, and that the Q30R substitution affects enzyme processing and secretion. The genome regions associated with hair features are enriched for signals of selection, consistent with proposals regarding the evolution of human hair.
Link: http://www.nature.com/ncomms/2016/160301/ncomms10815/full/ncomms10815.html
CANDELA paper on ear morphology published in Nature Communications:
'A genome-wide association study identifies multiple loci for variation in human ear morphology'
Summary from the article:
Here we report a genome-wide association study for non-pathological pinna morphology in over 5,000 Latin Americans. We find genome-wide significant association at seven genomic regions affecting: lobe size and attachment, folding of antihelix, helix rolling, ear protrusion and antitragus size (linear regression P values 2 × 10−8 to 3 × 10−14). Four traits are associated with a functional variant in the Ectodysplasin A receptor (EDAR) gene, a key regulator of embryonic skin appendage development. We confirm expression of Edar in the developing mouse ear and that Edar-deficient mice have an abnormally shaped pinna. Two traits are associated with SNPs in a region overlapping the T-Box Protein 15 (TBX15) gene, a major determinant of mouse skeletal development. Strongest association in this region is observed for SNP rs17023457 located in an evolutionarily conserved binding site for the transcription factor Cartilage paired-class homeoprotein 1 (CART1), and we confirm that rs17023457 alters in vitro binding of CART1.
Link: http://www.nature.com/ncomms/2015/150624/ncomms8500/full/ncomms8500.html
CANDELA paper on facial variation published in the American Journal of Physical Anthropology:
'Facial asymmetry and genetic ancestry in Latin American admixed populations'
Summary from the article:
Fluctuating and directional asymmetry are aspects of morphological variation widely used to infer environmental and genetic factors affecting facial phenotypes. However, the genetic basis and environmental determinants of both asymmetry types is far from being completely known. The analysis of facial asymmetries in admixed individuals can be of help to characterize the impact of a genome's heterozygosity on the developmental basis of both fluctuating and directional asymmetries. Here we characterize the association between genetic ancestry and individual asymmetry on a sample of Latin-American admixed populations. To do so, three-dimensional (3D) facial shape attributes were explored on a sample of 4,104 volunteers aged between 18 and 85 years. Individual ancestry and heterozygosity was estimated using more than 730,000 genome-wide markers. Multivariate techniques applied to geometric morphometric data were used to evaluate the magnitude and significance of directional and fluctuating asymmetry (FA), as well as correlations and multiple regressions aimed to estimate the relationship between facial FA scores and heterozygosity and a set of covariates. Results indicate that directional and FA are both significant, the former being the strongest expression of asymmetry in this sample. In addition, our analyses suggest that there are some specific patterns of facial asymmetries characterizing the different ancestry groups. Finally, we find that more heterozygous individuals exhibit lower levels of asymmetry. Our results highlight the importance of including ancestry-admixture estimators, especially when the analyses are aimed to compare levels of asymmetries on groups differing on socioeconomic levels, as a proxy to estimate developmental noise.
Link: http://onlinelibrary.wiley.com/doi/10.1002/ajpa.22688/abstract
CANDELA Introductory paper published in PLoS Genetics:
Summary from the article:
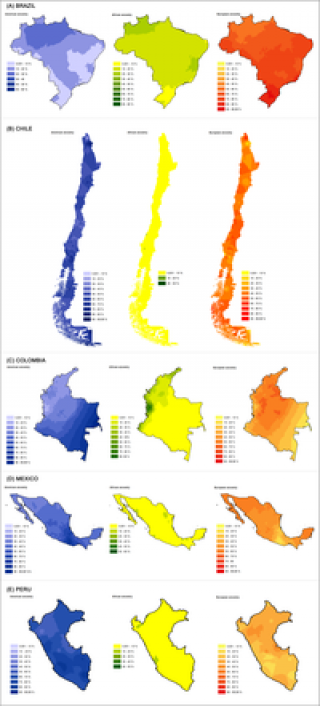
Latin America has a history of extensive mixing between Native Americans and people arriving from Europe and Africa. As a result, individuals in the region have a highly heterogeneous genetic background and show great variation in physical appearance. Latin America offers an excellent opportunity to examine the genetic basis of the differentiation in physical appearance between Africans, Europeans and Native Americans. The region is also an advantageous setting in which to examine the interplay of genetic, physical and social factors in relation to ethnic/racial self-perception. Here we present the most extensive analysis of genetic ancestry, physical diversity and self-perception of ancestry yet conducted in Latin America. We find significant geographic variation in ancestry across the region, this variation being consistent with demographic history and census information. We show that genetic ancestry impacts many aspects of physical appearance. We observe that self-perception is highly influenced by physical appearance, and that variation in physical appearance biases self-perception of ancestry relative to genetically estimated ancestry.
Link: http://www.plosgenetics.org/doi/pgen.1004572
Citation:
Ruiz-Linares A, Adhikari K, Acuña-Alonzo V, Quinto-Sanchez M, Jaramillo C, et al. (2014) Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals. PLoS Genet 10(9): e1004572. doi:10.1371/journal.pgen.1004572
CANDELA paper on human skin pigmentation published in PLoS ONE:
`Implications of the Admixture Process in Skin Color Molecular Assessment'
Summary from the article:
The understanding of the complex genotype-phenotype architecture of human pigmentation has clear implications for the evolutionary history of humans, as well as for medical and forensic practices. Although dozens of genes have previously been associated with human skin color, knowledge about this trait remains incomplete. In particular, studies focusing on populations outside the European-North American axis are rare, and, until now, admixed populations have seldom been considered. The present study was designed to help fill this gap. Our objective was to evaluate possible associations of 18 single nucleotide polymorphisms (SNPs), located within nine genes, and one pseudogene with the Melanin Index (MI) in two admixed Brazilian populations (Gaucho, N = 352; Baiano, N = 148) with different histories of geographic and ethnic colonization. Of the total sample, four markers were found to be significantly associated with skin color, but only two (SLC24A5 rs1426654, and SLC45A2 rs16891982) were consistently associated with MI in both samples (Gaucho and Baiano). Therefore, only these 2 SNPs should be preliminarily considered to have forensic significance because they consistently showed the association independently of the admixture level of the populations studied. We do not discard that the other two markers (HERC2 rs1129038 and TYR rs1126809) might be also relevant to admixed samples, but additional studies are necessary to confirm the real importance of these markers for skin pigmentation. Finally, our study shows associations of some SNPs with MI in a modern Brazilian admixed sample, with possible applications in forensic genetics. Some classical genetic markers in Euro-North American populations are not associated with MI in our sample. Our results point out the relevance of considering population differences in selecting an appropriate set of SNPs as phenotype predictors in forensic practice.
Link: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0096886
Citation:
Cerqueira CCSd, Hünemeier T, Gomez-Valdés J, Ramallo V, Volasko-Krause CD, et al. (2014) Implications of the Admixture Process in Skin Color Molecular Assessment. PLoS ONE 9(5): e96886. doi:10.1371/journal.pone.0096886
CANDELA paper on genetic ancestry distribution in Chile:
Summary from the article:
BACKGROUND:
The geographical distribution of genes plays a key role in genetic epidemiology. The Chilean population has three major stem groups (Native American, European and African).
AIM:
To estimate the regional rate of American, European and African admixture of the Chilean population.
SUBJECTS AND METHODS:
Forty single nucleotide polymorphisms (SNP´s) which exhibit substantially different frequencies between Amerindian populations (ancestry-informative markers or AIM´s), were genotyped in a sample of 923 Chilean participants to estimate individual genetic ancestry.
RESULTS:
The American, European and African individual average admixture estimates for the 15 Chilean Regions were relatively homogeneous and not statistically different. However, higher American components were found in northern and southern Chile and higher European components were found in central Chile. A negative correlation between African admixture and latitude was observed. On the average, American and European genetic contributions were similar and significantly higher than the African contribution. Weighted mean American, European and African genetic contributions of 44.34% ± 3.9%, 51.85% ± 5.44% and 3.81% ± 0.45%, were estimated. Fifty two percent of subjects harbor African genes. Individuals with Aymara and Mapuche surnames have an American admixture of 58.64% and 68.33%, respectively.
CONCLUSIONS:
Half of the Chilean population harbors African genes. Participants with Aymara and Mapuche surnames had a higher American genetic contribution than the general Chilean population. These results confirm the usefulness of surnames as a first approximation to determine genetic ancestry.
Link: http://www.scielo.cl/scielo.php?script=sci_arttext&pid=S0034-98872014000300001&lng=en&nrm=iso&tlng=en
Citation:
Fuentes, Macarena et al. Gene geography of Chile: Regional distribution of American, European and African genetic contributions. Rev. méd. Chile. 2014, vol.142, n.3, pp. 281-289. http://dx.doi.org/10.4067/S0034-98872014000300001.
 Close
Close

