Electron/Proton Transfer of Respiratory Chain Enzymes
Our basic research focuses on biological electron and proton transfer processes in electron transfer chain, particularly those of the respiratory chain. Major techniques are vibrational infrared (IR) and UV/visible spectroscopies, together with a wide range of other biophysical and biochemical methods. Particular areas of experimental interest are cytochrome c oxidase and related enzymes such as peroxidases.
Cytochrome c oxidase overview
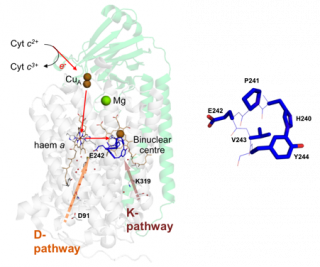
4 cyt c2+ + O2 + 8 H+ (matrix) → 4 cyt c3+ + 2 H2O + 4 H+ (intermembrane space) Structure of bovine complex IV (subunits I and II) and its prosthetic groups (taken from Rich, P.R. and Maréchal, A. (2010) The mitochondrial respiratory chain. Essays in Biochemistry, 47, 1-23. Cytochrome c oxidase (CcO) is a mitochondrial membrane protein that catalyses the reduction of oxygen to water with electrons from ferrocytochrome c in the intermembrane space and four protons from the mitochondrial matrix. The reaction is coupled to the translocation of four additional protons across the inner mitochondrial membrane. These reactions generate a protonmotive force which is used to drive endergonic processes such as ATP synthesis. The oxygen reduction reaction itself takes place in a binuclear centre that is composed of haem a3 and CuB and protons are moved through hydrophilic channels in the protein structure.
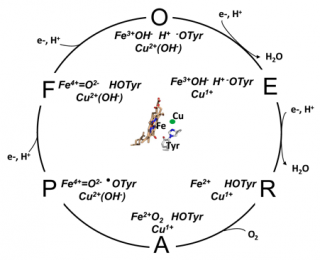
Cytochrome c oxidase reaction cycle
The oxygen reduction reaction cycle of cytochrome c oxidase (taken from Rich, P.R. and Maréchal, A. (2012) Respiratory Chains: Structures, Mechanisms and Energy Coupling. In: Edward H. Egelman, editor: Comprehensive Biophysics, Vol 8, Chapter 6, Bioenergetics (Stuart Ferguson, ed.), pp. 72-93, Academic Press, Oxford). The oxidized (O) state is drawn with hydroxide ligands on both Cu
B and haem a
3, an anionic tyrosinate form of the covalent histidine-tyrosine and a further proton shared between haem hydroxide and tyrosinate.
Each of the four electron transfers into the BNC is accompanied by uptake of a charge-compensating substrate proton. The O→E and E→R steps are both assumed to result in loss of an H2O, leaving the metals free to ligate O2 and form the oxyferrous species A. A decomposes in microseconds to P, breaking the O=O bond to form a ferryl heme (Fe4+=O2-), CuB2+OH- and a neutral tyrosine radical (●OTyr). The radical is re-reduced by the third electron/proton transfer in the P→F step. The final, fourth, electron/proton transfer in F→O reduces the ferryl haem and so regenerates the starting O state configuration. The four electron/proton transfer steps are likely to be linked to common proton translocation mechanism.
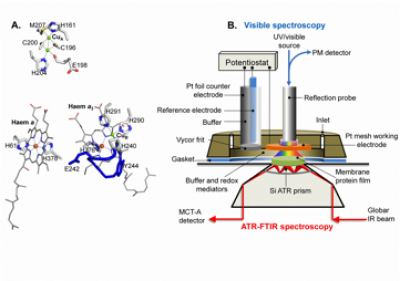
Example of electrochemistry coupled ATR-FTIR spectroscopy
CcO metals centres and their ligands and electrochemical ATR-FTIR/UV-visible sample cell (taken from Dodia, R. et al. (2013) IR signatures of the metal centres of bovine cytochrome c oxidase: assignments and redox-linkage. Biochemical Society Transactions, 41, 5, 1242-1248). Functionally significant redox-linked structural changes of cytochrome c oxidase can be studied using electrochemistry coupled ATR-FTIR spectroscopy as shown above. The electrochemical cell is designed to also facilitate in situ UV/visible spectroscopy measurements for monitoring the reaction and the stability of the sample. In addition to the example shown above, we have developed devices that allow diverse reactions to be induced using perfusion/dialysis methods that allow introduction and removal of reactants, such as ligands, metal ions, redox mediators.
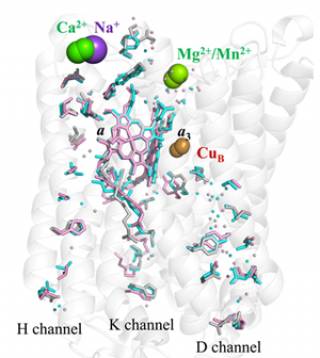
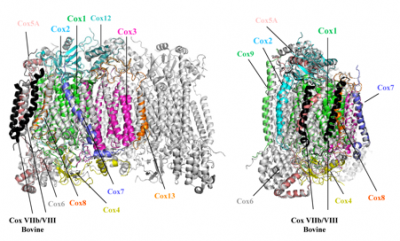
Yeast vs. Bovine cytochrome c oxidases
Superposition of homology models of the 11 subunits of yeast CcO on the dimeric bovine CcO structure (taken from Maréchal, A. et al. (2012) Yeast cytochrome
c oxidase: A model system to study mitochondrial forms of the haem-copper oxidase superfamily. Biochimica et Biophysica Acta, 1817 (4), 620-8). Mitochondrial cytochrome c oxidase is a large multimeric protein composed of 13 (bovine) or 11 (yeast) different subunits. Three of these subunits are encoded by the mitochondrial genome and the remainder by the nuclear genome. Bacterial homologues are simpler, with a functional core composed of the same three subunits that are mitochondrially-encoded in eukaryotes. A structural model of yeast cytochrome
c oxidase is shown in colour that was predicted by homology modelling with the known bovine cytochrome c oxidase structure (which is dimeric in the crystal form) shown in grey. The yeast cytochrome c oxidase is a good model system to investigate the coupling of proton and electron transfer processes, as well as the role of the possible proton translocation pathways, since we are able to generate mutant forms of this enzyme.







 Close
Close