Molecular genetic techniques, including whole-genome sequencing, are providing novel ways to track the spread of infections and the evolution of microorganisms.
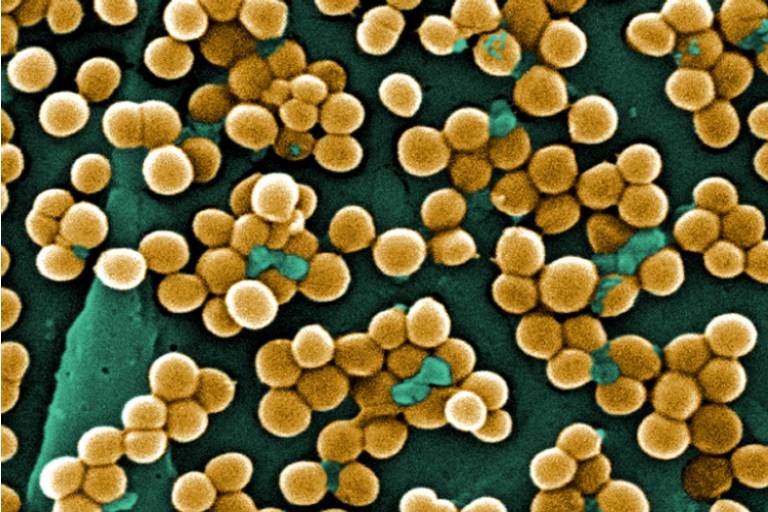
Genomic approaches are increasingly used to categorise and identify strains of bacteria. Furthermore, DNA sequencing technologies are sufficiently advanced that whole-genome sequencing of pathogens is near-routine.
One use of this kind of information is in the characterisation of transmission chains in clinical facilities such as hospitals. A comparison of genome sequences can reveal whether samples are related, and hence are part of the same outbreak. Family trees of related sequences can also reveal the likely paths of transmission. Such information can be critical to effective infection control.
On a wider scale, sequencing of samples collected at difference places and/or different points in time can enable the evolutionary history of bacterial strains or species to be reconstructed. Such work can provide vital insight into how antibiotic-resistant strains are evolving and being disseminated regionally and globally.
 Close
Close

