UCL Genetics Institute
Welcome
Our research is both fundamental and applied, and we believe there does not need to be a divide between the two. Important Science is by definition fundamental, and is also likely to have downstream applications.
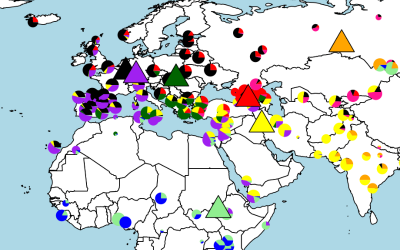
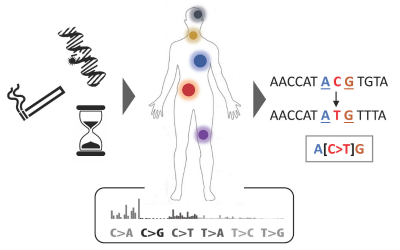
We develop computational tools and apply them to large genetic datasets to address questions ranging from the origin and evolution of anatomically modern humans, the genetic determinants of various human diseases, improvement of crop varieties to antibiotic resistance in bacteria.

About Us
News
Research themes
Study


Read the Paper

Read the Paper

Read the Paper

Read the Paper

Read the Paper

Read the Paper

 Close
Close