In vitro models to study lung cancer and airway regeneration
As part of our airway epithelial regeneration work we have developed methods to expand human airway epithelial cells from small endobronchial biopsy samples. After expansion, we differentiate these towards the mature airway cell types, including ciliated and mucosecretory cells, using either air-liquid interface or 3D organoid (or 'tracheosphere') assays (shown in the image below). You can read more about our group's work on the expansion of human epithelial stem cells for tissue engineering here (Butler et al. 2016).
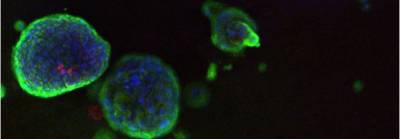 |
| Human airway organoids |
We are investigating the relationship between cancer cells and the stroma using spheroids embedded in tunable stiffness collagen matrices, containing stromal cells or PDMS gel. There is compelling evidence for the role of stroma and its physical interaction with epithelial cells in the acts of metastasis. Stiffer tissues and increased extracellular matrix (ECM) stiffness have been shown to affect several processes associated with tumour progression. The images below show three dimensional cultures of normal human bronchial epithelial cells (left) and lung squamous carcinoma cells (right), in co-culture with fibroblast cells and in a collagen matrix.
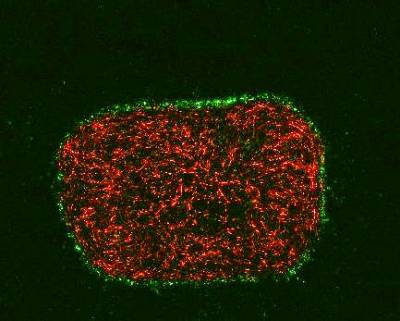 | 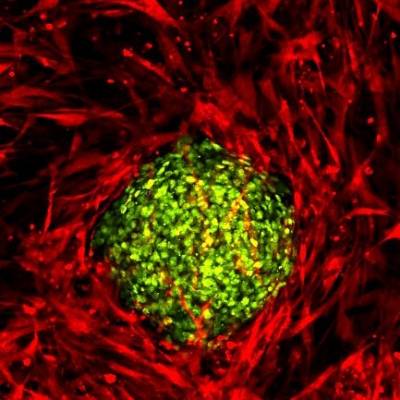 |
Wild type fibroblasts stably expressing a red fluorescent protein embedded into collagen gel. HBEC cells expressing nuclear GFP were seeded on top of the gel and organized accordingly. | Wildtype fibroblasts stably expressing a red fluorescent protein embedded into collagen gel. Spheroids of lung cancer cell line H2170 expressing nuclear GFP were seeded on top of the gel. |
Our group is also interested in the signalling changes taking place during cellular transformation. We use both lentiviral and CRISPR/Cas9 methods of genetic manipulation to study the effects of specifically changing the expression of genes thought to be critical in lung cancer development. The left image below shows normal lung epithelial cells, expressing tumour suppressor protein p53, stained in white. This compares to the absence of p53 in the cells shown in the right image, which underwent a CRISPR/Cas9 targeted deletion of p53.
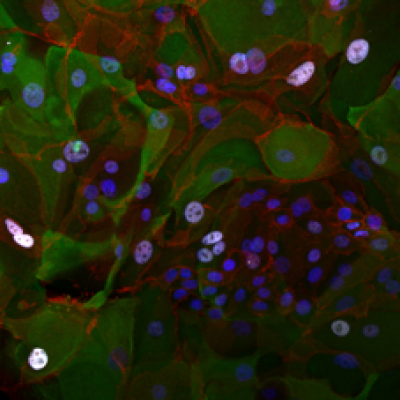 | 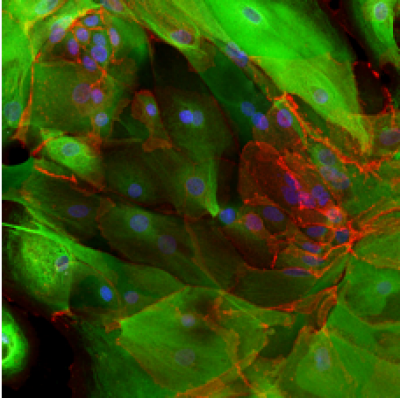 |
Control cells | p53-negative cells |
Bioinformatic and computational expertise
Modern sequencing techniques generate vast volumes of data. For many years, our team has been applying these techniques to the very earliest pre-cancerous changes in the lung, and even to biopsies of healthy lung tissue to understand how lung cancer forms. Our group has in-house expertise in bioinformatics, the field developed to extract meaning from these data. We have published the most comprehensive molecular analysis of preinvasive lung cancer to date, studying the genomic, transcriptomic and epigenetic landscape of pre-cancer to isolate very early cancerous changes, which could one day provide novel therapeutic targets (Teixeira et al. 2019).
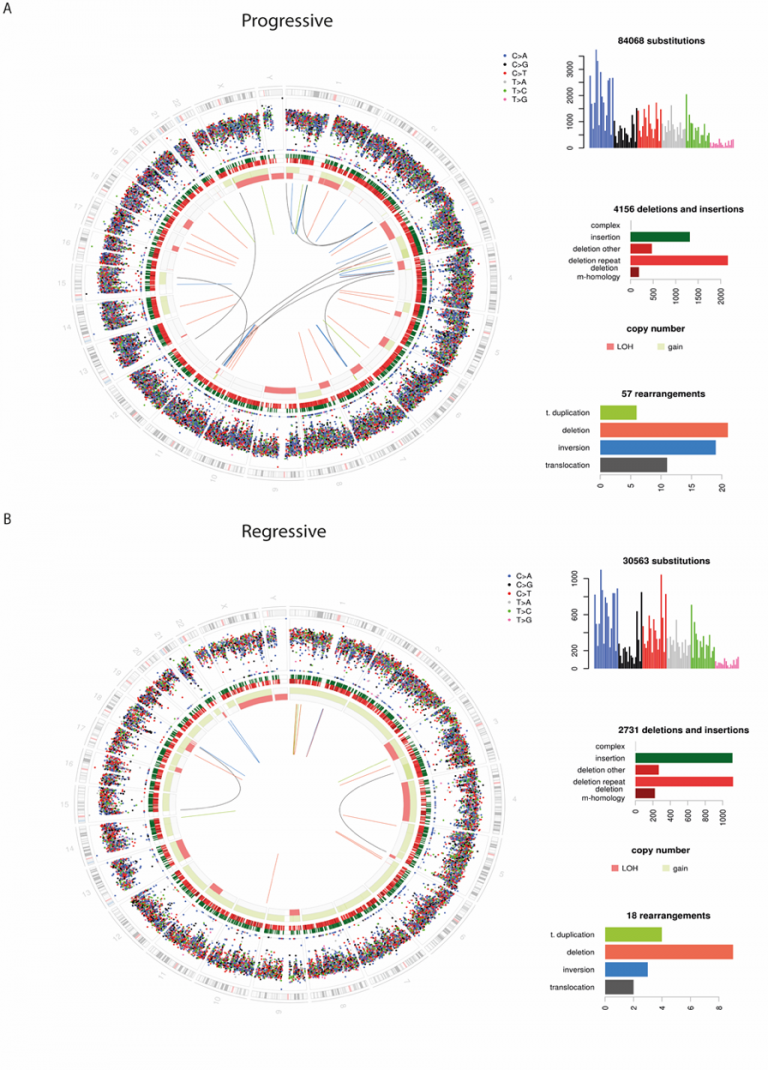
Whilst we have published the most interesting findings from this vast dataset, we are continually gaining new insights as new analysis techniques become available. Furthermore we are continually acquiring new and exciting datasets to better understand this field. Through a large international collaboration we will be sequencing many more similar pre-cancerous lesions, and applying novel techniques such as single cell transcriptomics. New sample collection pipelines, for example from lung cancer screening studies, are opening up new data-driven projects.
Our computational capacity is underpinned by UCL’s world-class infrastructure, alongside close collaboration with the Sanger Institute and the Francis Crick Institute. We work closely with these institutions to leverage the wealth of bioinformatic tools developed in advanced cancer and apply these to the new frontier of early carcinogenesis. By understanding these early steps in cancer formation we hope to develop new strategies for early detection, treatment and interception.
Infrared (IR) spectroscopy
Infrared (IR) spectroscopy is an optical technology that can be used to detect differences between diseased and healthy tissue based on their biochemical profiles.
We are using IR spectroscopy to identify a buccal spectral biomarker to improve the early diagnosis of lung cancer. This research is funded by the Roy Castle Foundation and performed in collaboration with DynaMx Medical and supported by Diamond Light Source, Harwell.
This project uses two IR techniques, Fourier Transform Infrared Spectroscopy (FTIR) and attenuated total reflection (ATR)-FTIR. In FTIR, light with a wide spectrum of wavelengths is passed through a sample. The resultant spectra generated creates a ‘biochemical fingerprint’ sensitive to changes in a wide range of compounds including carbohydrates, lipids, proteins and DNA. In ATR-FTIR, light is totally internally reflected through use of an optically configured IR transmitting prism. ATR-FTIR can be used on samples taken directly from the patient and placed on the prism with no need for preparation and as such has potential for use in a point-of-care device.
We have optimised our sample preparation techniques and are looking to use IR spectroscopy to establish algorithms to validate an infrared spectral biomarker of lung cancer risk.
 Close
Close

