Jason Mercer tells the story behind the paper
Jason Mercer and co-author Bernd Wollscheid (ETH Zurich) tell the story behind their recent paper in Nature Microbiology: Vaccinia, mass spectrometry and electron microscopy: It takes three to tango.
Find the research article here.
Abstract from the article:
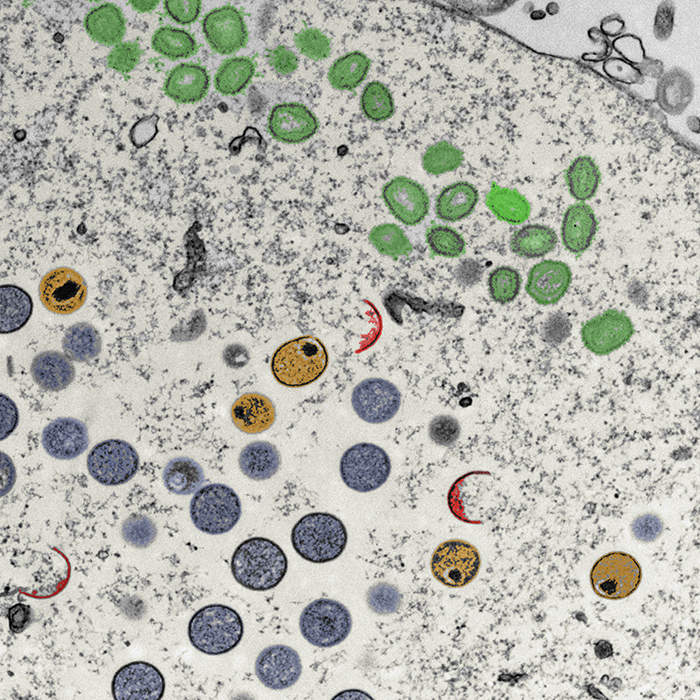
To orchestrate context-dependent signalling programmes, poxviruses encode two dual-specificity enzymes, the F10 kinase and the H1 phosphatase. These signalling mediators are essential for poxvirus production, yet their substrate profiles and systems-level functions remain enigmatic. Using a phosphoproteomic screen of cells infected with wild-type, F10 and H1 mutant vaccinia viruses, we systematically defined the viral signalling network controlled by these enzymes. Quantitative cross-comparison revealed 33 F10 and/or H1 phosphosites within 17 viral proteins. Using this proteotype dataset to inform genotype–phenotype relationships, we found that H1-deficient virions harbour a hidden hypercleavage phenotype driven by reversible phosphory-lation of the virus protease I7 (S134). Quantitative phosphoproteomic profiling further revealed that the phosphorylation-dependent activity of the viral early transcription factor, A7 (Y367), underlies the transcription-deficient phenotype of H1 mutant virions. Together, these results highlight the utility of combining quantitative proteotype screens with mutant viruses to uncover proteotype–phenotype–genotype relationships that are masked by classical genetic studies.
Find out about the Mercer Lab.
 Close
Close