Neurogenesis and Brain Cancer
Group Leader: Professor Simona Parrinello
Professor of Neuro-oncology and Co-lead of the UCL Cancer Research UK Brain Tumour Centre of Excellence
Research
Neural stem cells persist in restricted neurogenic regions of the adult brain where they continuously generate new neurons and glia. Neural stem cell behaviour is controlled by a complex interplay between transcriptional programs and extrinsic cues from the microenvironment, or niche, in which the stem cells reside.
Glioblastoma, the most common and aggressive type of adult brain tumour, also contains subpopulations of cancer cells with neural stem cell properties, termed glioma stem cells or GSCs. GSCs drive tumour growth, give rise to more differentiated tumour bulk cells and underlie resistance to therapy. Increasing evidence suggests that GSCs promote tumourigenesis by hijacking the transcriptional and signalling networks that control normal neurogenesis. Studying NSC biology and its links to GSC malignancy will therefore increase understanding of this devastating disease and lead to the development of more effective therapeutic strategies.
The main interests of my group are to understand the cellular and molecular mechanisms by which the niche controls normal NSC function and to determine how these mechanisms become deregulated in GSCs to drive tumour growth and invasion. To address these questions, we use a combination of experimental approaches, including cell biology, gene editing, proteomics, high throughput sequencing and advanced intravital microscopy in mouse models and patient-derived cells.
Current projects in the lab focus on three complementary areas (Figure 1):
- The role of the niche in controlling the activation of quiescent neural stem cells
- The molecular basis of oncogene and injury-induced dedifferentiation in adult glia
- The microenvironmental signals that maintain GSC quiescence and drive glioblastoma invasion
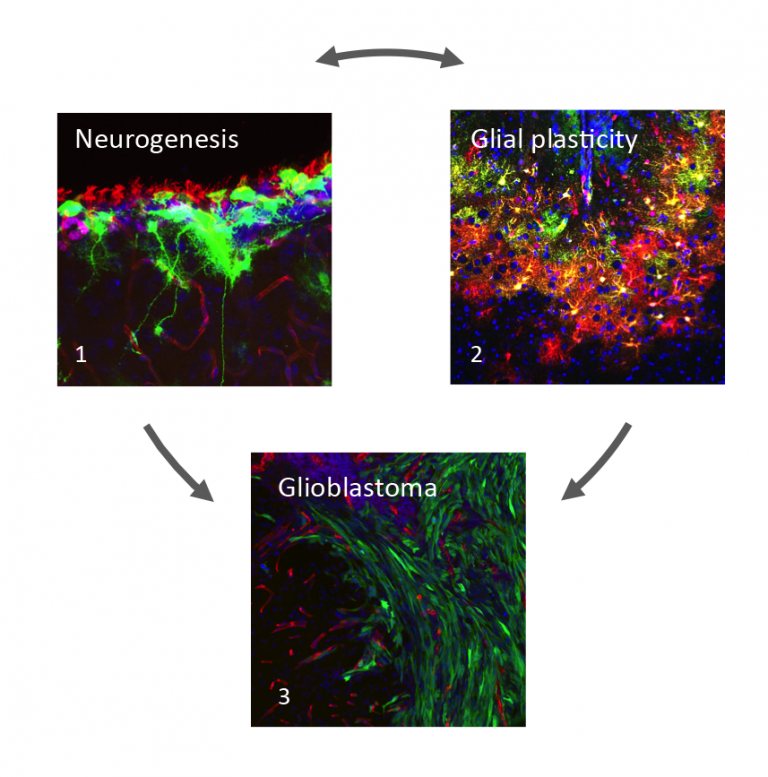
Figure 1
1.Neural stem cells (green) contacting apical ciliated ependymal cells and basal blood vessels (red) in the subventricular zone niche.
2.Reactive astrocytes (green and red) surrounding a stab wound injury.
3.Tumour cells (green) invading into the normal brain parenchyma along blood vessels (red).
We welcome informal inquiries, please contact Simona Parrinello s.parrinello@ucl.ac.uk if you would like to work with us.
Selected publications
- Brooks LJ, Clements MP, Burden JJ, Kocher D, Richards L, Devesa SC, Zakka L,Woodberry M, Ellis M, Jaunmuktane Z, Brandner S, Morrison G, Pollard SM, Dirks PB, Marguerat S, Parrinello S. The white matter is a pro-differentiative niche for glioblastoma. Nat Commun. 2021 Apr 12; 12(1):2184. doi: 10.1038/s41467-021-22225-w. PMID: 33846316; PMCID: PMC8042097
- Garcia-Diaz C, Mereu E, Clements MP, Poysti A, Galvez-Cancino F, Castillo SP, Courtot L, Ruiz S, Roncaroli F, Yuan Y, Quezada SA, Heyn H, Parrinello S. Glioblastoma cell fate is differentially regulated by the microenvironments of the tumour bulk and infiltrative margin. bioRxiv. 2021 Jun.
- Amodeo V, Davies T, Martinez-Segura A, Clements MP, Simpson Ragdale H, Bailey A, Silva Dos Santos M, MacRae JI, Mokochinski J, Kramer H, Garcia-Diaz C, Gould AP, Marguerat S, Parrinello S. Diet supresses tumour initiation by maintaining quiescene of mutation-bearing neural stem cells. bioRxiv. 2022 March.
- Brooks LJ, Parrinello S. Vascular regulation of glioma stem-like cells: a balancing act. Curr Opin Neurobiol. 2017 Dec;47:8-15.
- Clements MP, Byrne E, Camarillo Guerrero LF, Cattin AL, Zakka L, Ashraf A, Burden JJ, Khadayate S, Lloyd AC, Marguerat S, Parrinello S. The Wound Microenvironment Reprograms Schwann Cells to Invasive Mesenchymal-like Cells to Drive Peripheral Nerve Regeneration. Neuron. 2017 Sep 27;96(1):98-114.e7
- Krusche B, Ottone C, Clements MP, Johnstone ER, Goetsch K, Lieven H, Mota SG, Singh P, Khadayate S, Ashraf A, Davies T, Pollard SM, De Paola V, Roncaroli F, Martinez-Torrecuadrada J, Bertone P, Parrinello S. EphrinB2 drives perivascular invasion and proliferation of glioblastoma stem-like cells. Elife. 2016 Jun 28;5. pii: e14845.
- Ottone C, Parrinello S. Multifaceted control of adult SVZ neurogenesis by the vascular niche. Cell Cycle. 2015;14(14):2222-5.
- Ottone C, Krusche B, Whitby A, Clements M, Quadrato G, Pitulescu ME, Adams RH, Parrinello S. Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells. Nat Cell Biol. 2014 Nov;16(11):1045-56.
- Parrinello S, Napoli I, Ribeiro S, Wingfield Digby P, Fedorova M, Parkinson DB, Doddrell RD, Nakayama M, Adams RH, Lloyd AC. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 2010 Oct 1;143(1):145-55.
 Close
Close





