Project leader: Emanuele Libertini
Our current computational analysis approaches entail developing methods for the integration of epigenomic next generation sequencing data with genomic, transcriptomic and other types of datasets. The primary focus is whole genome bisulfite sequencing, yielding a high resolution genome-wide profile of DNA methylation. Our work so far has benchmarked the potential of this base resolution technique by assessing the existing computational tools, the effects of smoothing, developing new tools (COMETgazer) and performing feature-specific saturation analyses. These highlighted the effects of coverage on biological signature extraction.
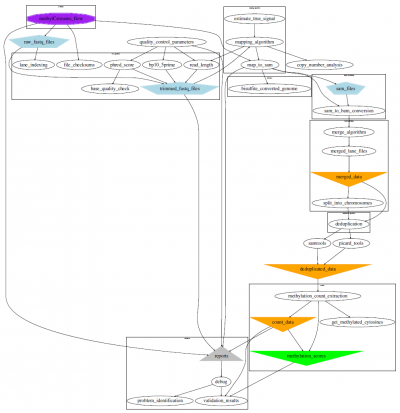
Our experience with these data brought about the assembly of a WGBS workflow for full analysis including processing compartments for quality control to alignment and extraction and on to the analysis of differentially methylated positions and regions, as well as the determination of blocks of co-methylation (COMETs) with our COMETgazer algorithm, which exploits DNA methylation oscillations.
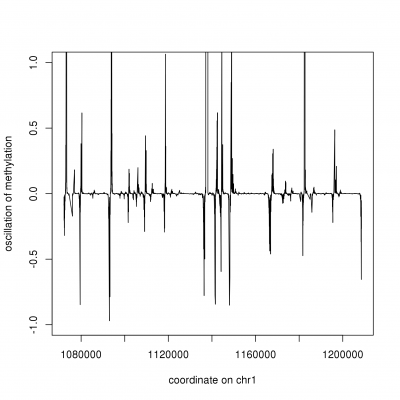
CpG methylation can be modelled as an harmonic, where oscillatory patterns are used to segment the methylome
Our approach to differential methylation analysis is based on our experience of the effect of coverage, utilizing point-wise, regional or COMET inference depending on sequencing depth and data resolution. We use the COMET approach for low-resolution analyses as a means to summarize methylation over large DNA stretches and compute differential methylation by assessing the fragmentation of the methylome. Our COMET work is currently in review with Nature Biotechnology.
Our data integration approaches focus on the use of COMET profiles for assessing a potential relationship with haplotype blocks and for the prediction of genomic enhancers. We are developing methods for harvesting oscillations in DNA methylation for epigenomic signature analysis.
We also have experience with the analysis of other next generation sequencing dataset types including RNA-seq, ChIP-Seq and MeDIPSeq, and the integration of these with transcriptomic datasets. A previous lab member (Gareth Wilson) developed a MeDIPSeq analysis wrapper (the MeDUSA pipeline) for the analysis of MeDIPSeq data (Wilson et al., 2012). The focus of which is to locate differentially methylated regions between cohorts.
Our group is involved in the UK contribution to the Personal Genome Project which aims at creating UK genome, health and trait data integration with epigenomics in a open data, open access framework.
 Close
Close